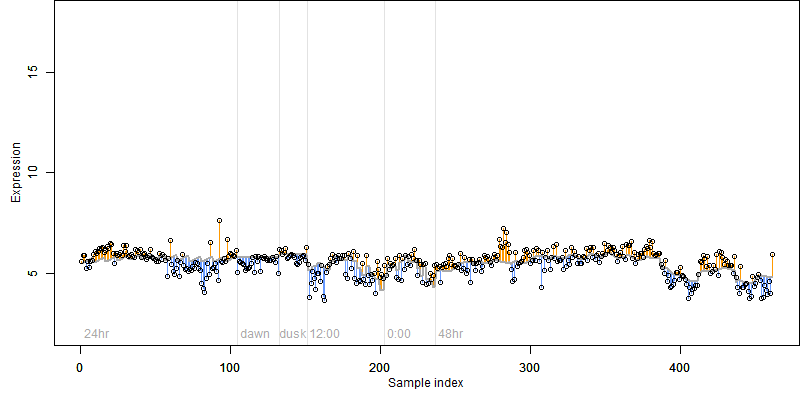
Os12g0500700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Vacuolar sorting protein-like; embryogenesis protein H beta 58-like protein.

FiT-DB / Search/ Help/ Sample detail
|
Os12g0500700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Vacuolar sorting protein-like; embryogenesis protein H beta 58-like protein.
|
|
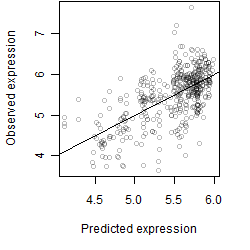
log2(Expression) ~ Norm(μ, σ2)
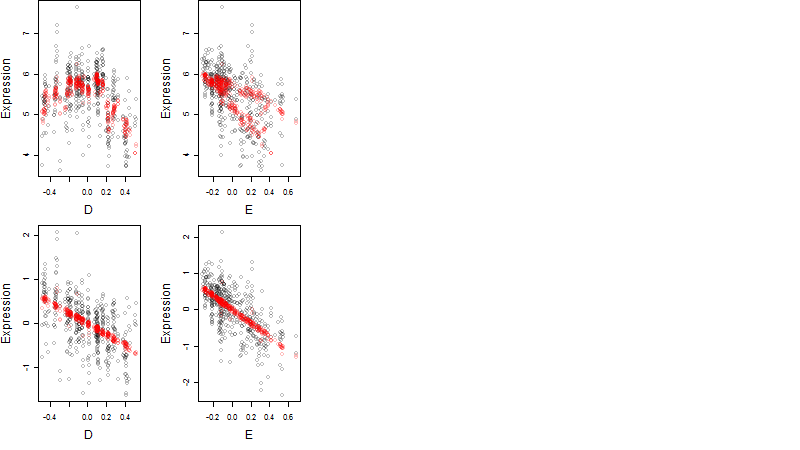
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

Dependence on each variable
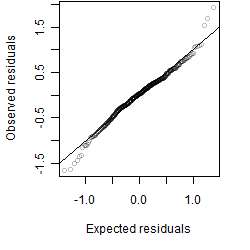
Residual plot
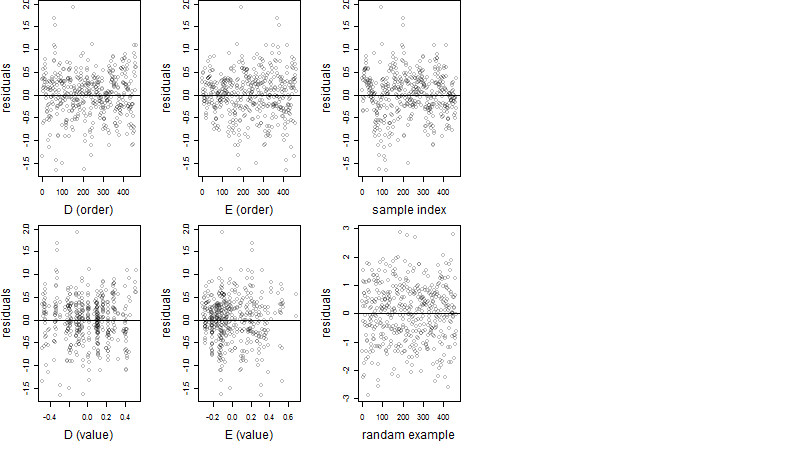
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 103.5 | 0.478 | 5.51 | -1.08 | 0.235 | -1.88 | 1.13 | -0.628 | -0.138 | 15.4 | 31.6 | 1514 | -- | -- |
| 103.84 | 0.475 | 5.53 | -1.21 | 0.239 | -1.88 | 1.12 | -- | -0.166 | 15.4 | 32.14 | 1517 | -- | -- |
| 107.4 | 0.483 | 5.51 | -1.06 | 0.189 | -1.88 | -- | -0.596 | -0.133 | 15.3 | 31.67 | 1627 | -- | -- |
| 107.61 | 0.483 | 5.52 | -1.23 | 0.175 | -1.87 | -- | -- | -0.151 | 14.9 | 32.93 | 1907 | -- | -- |
| 108.72 | 0.486 | 5.51 | -1.08 | -- | -1.88 | -- | -0.631 | -0.137 | -- | 31.56 | 1788 | -- | -- |
| 162.94 | 0.598 | 5.45 | -0.608 | 0.186 | -- | 1.07 | -- | 0.206 | 15.6 | -- | -- | -- | -- |
| 166.45 | 0.604 | 5.44 | -0.591 | 0.193 | -- | -- | -- | 0.212 | 15.9 | -- | -- | -- | -- |
| 109.07 | 0.486 | 5.52 | -1.21 | -- | -1.87 | -- | -- | -0.152 | -- | 32.17 | 1788 | -- | -- |
| 139.84 | 0.551 | 5.47 | -- | 0.345 | -1.38 | -- | -- | 0.0692 | 15 | 33.1 | 1250 | -- | -- |
| 168.5 | 0.607 | 5.44 | -0.599 | -- | -- | -- | -- | 0.207 | -- | -- | -- | -- | -- |
| 175.65 | 0.62 | 5.43 | -- | 0.205 | -- | -- | -- | 0.278 | 15.9 | -- | -- | -- | -- |
| 141.25 | 0.554 | 5.47 | -- | -- | -1.44 | -- | -- | 0.0671 | -- | 31.5 | 406 | -- | -- |
| 177.96 | 0.623 | 5.43 | -- | -- | -- | -- | -- | 0.273 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance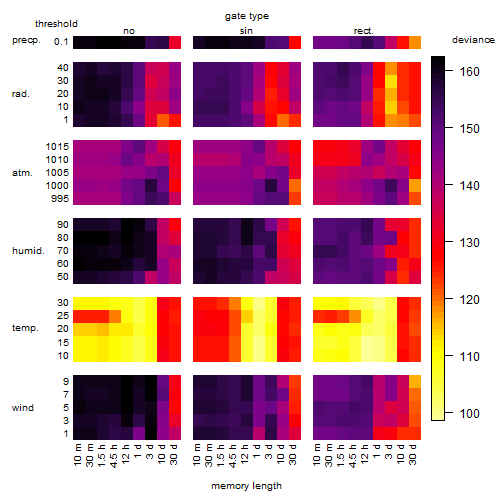
|
Histogram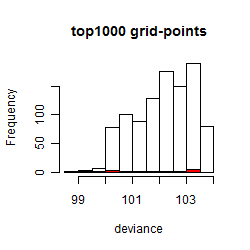 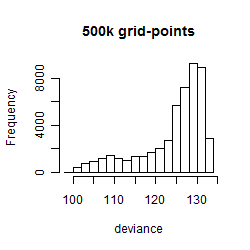
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 98.65 | temperature | 10 | 720 | > th | dose dependent | rect. | 21 | 18 |
| 3 | 99.35 | temperature | 15 | 720 | > th | dose dependent | rect. | 19 | 20 |
| 8 | 99.98 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 15 |
| 12 | 100.05 | temperature | 15 | 1440 | > th | dose dependent | rect. | 18 | 14 |
| 33 | 100.25 | temperature | 15 | 1440 | > th | dose dependent | sin | 10 | NA |
| 41 | 100.28 | temperature | 30 | 1440 | < th | dose dependent | sin | 11 | NA |
| 176 | 100.94 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 8 |
| 211 | 101.12 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 21 |
| 272 | 101.48 | temperature | 30 | 1440 | < th | dose dependent | rect. | 3 | 5 |
| 341 | 101.77 | temperature | 10 | 1440 | > th | dose dependent | rect. | 5 | 2 |
| 367 | 101.86 | temperature | 30 | 1440 | < th | dose dependent | rect. | 5 | 2 |
| 467 | 102.22 | temperature | 15 | 1440 | > th | dose dependent | rect. | 23 | 1 |
| 477 | 102.24 | temperature | 30 | 1440 | < th | dose dependent | rect. | 23 | 1 |
| 730 | 102.99 | temperature | 30 | 720 | < th | dose dependent | rect. | 24 | 21 |
| 747 | 103.07 | temperature | 30 | 1440 | < th | dose dependent | rect. | 3 | 1 |
| 767 | 103.11 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 1 |
| 774 | 103.13 | temperature | 15 | 1440 | > th | dose dependent | rect. | 1 | 1 |
| 910 | 103.49 | temperature | 15 | 1440 | > th | dose dependent | rect. | 3 | 1 |