Os11g0507400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.
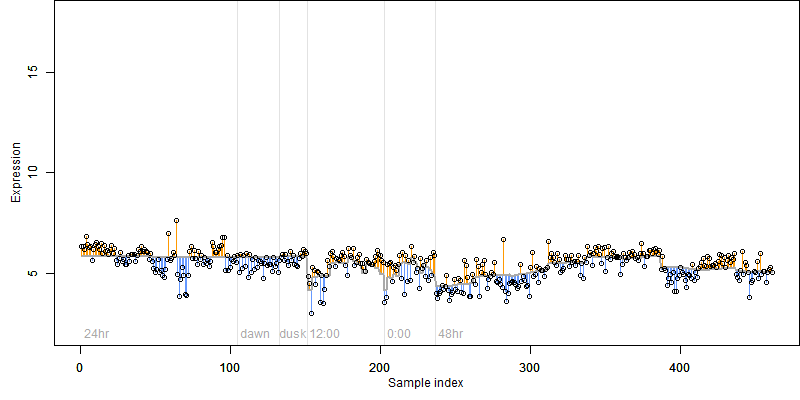

FiT-DB / Search/ Help/ Sample detail
|
Os11g0507400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
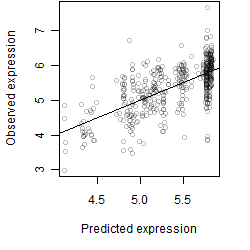
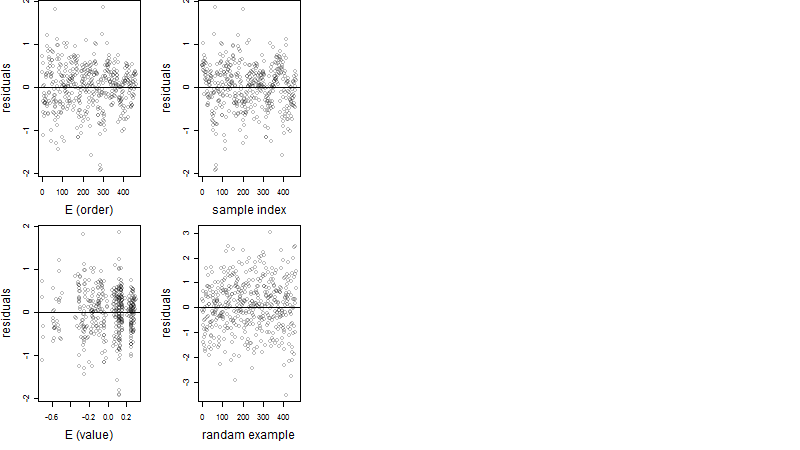
Dependence on each variable

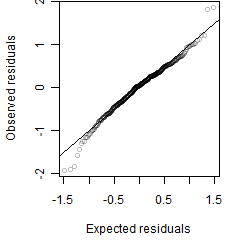
Residual plot
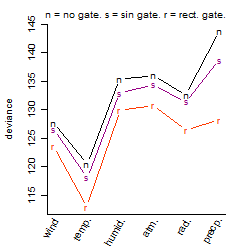
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 119.66 | 0.514 | 5.33 | 0.189 | 0.164 | 1.5 | 0.478 | -0.617 | 0.309 | 13.3 | 10.44 | 4729 | -- | -- |
| 119.28 | 0.509 | 5.32 | 0.289 | 0.169 | 1.58 | 0.546 | -- | 0.3 | 13.9 | 11.57 | 5838 | -- | -- |
| 120.07 | 0.51 | 5.34 | 0.155 | 0.16 | 1.54 | -- | -0.656 | 0.263 | 13 | 11.16 | 5839 | -- | -- |
| 120.46 | 0.511 | 5.32 | 0.299 | 0.166 | 1.58 | -- | -- | 0.304 | 13.4 | 10.39 | 5842 | -- | -- |
| 121.57 | 0.514 | 5.35 | 0.138 | -- | 1.52 | -- | -0.723 | 0.253 | -- | 9.918 | 5970 | -- | -- |
| 161.39 | 0.596 | 5.24 | 1.04 | 0.125 | -- | 0.5 | -- | 0.647 | 14.2 | -- | -- | -- | -- |
| 162.23 | 0.596 | 5.24 | 1.06 | 0.129 | -- | -- | -- | 0.65 | 12.9 | -- | -- | -- | -- |
| 122.01 | 0.514 | 5.32 | 0.295 | -- | 1.57 | -- | -- | 0.296 | -- | 10.3 | 5975 | -- | -- |
| 122.16 | 0.515 | 5.33 | -- | 0.158 | 1.74 | -- | -- | 0.238 | 12.9 | 8.488 | 5942 | -- | -- |
| 163.28 | 0.597 | 5.24 | 1.05 | -- | -- | -- | -- | 0.647 | -- | -- | -- | -- | -- |
| 191.77 | 0.648 | 5.27 | -- | 0.113 | -- | -- | -- | 0.532 | 12.5 | -- | -- | -- | -- |
| 123.68 | 0.518 | 5.34 | -- | -- | 1.73 | -- | -- | 0.235 | -- | 8.55 | 5981 | -- | -- |
| 192.57 | 0.648 | 5.27 | -- | -- | -- | -- | -- | 0.53 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance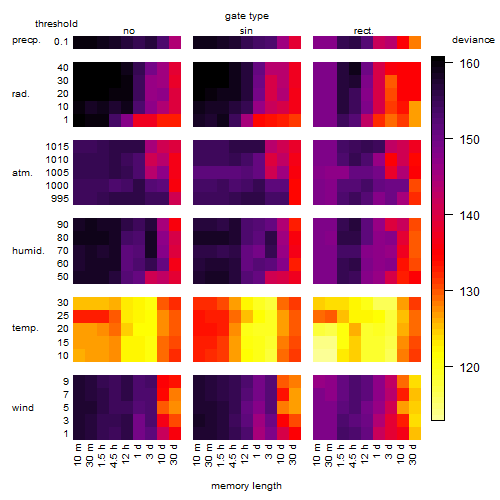
|
Histogram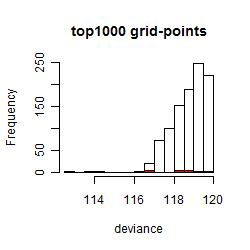 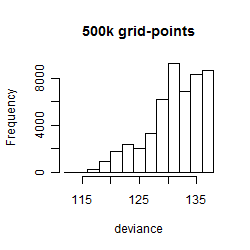
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 112.85 | temperature | 10 | 30 | > th | dose dependent | rect. | 23 | 23 |
| 5 | 116.48 | temperature | 30 | 4320 | < th | dose dependent | rect. | 23 | 1 |
| 6 | 116.55 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 2 |
| 7 | 116.57 | temperature | 10 | 4320 | > th | dose dependent | rect. | 23 | 3 |
| 8 | 116.57 | temperature | 30 | 4320 | < th | dose dependent | rect. | 23 | 3 |
| 11 | 116.74 | temperature | 10 | 4320 | > th | dose dependent | rect. | 1 | 1 |
| 13 | 116.79 | temperature | 30 | 4320 | < th | dose dependent | rect. | 1 | 1 |
| 200 | 118.05 | temperature | 10 | 4320 | > th | dose dependent | sin | 12 | NA |
| 219 | 118.10 | temperature | 30 | 4320 | < th | dose dependent | sin | 12 | NA |
| 259 | 118.26 | temperature | 15 | 1440 | > th | dose dependent | rect. | 18 | 1 |
| 307 | 118.39 | temperature | 10 | 720 | > th | dose dependent | rect. | 13 | 22 |
| 404 | 118.66 | temperature | 30 | 4320 | < th | dose dependent | rect. | 3 | 2 |
| 430 | 118.73 | temperature | 10 | 4320 | > th | dose dependent | rect. | 3 | 2 |
| 432 | 118.73 | temperature | 20 | 720 | > th | dose dependent | rect. | 19 | 16 |
| 433 | 118.74 | temperature | 20 | 4320 | > th | dose dependent | rect. | 3 | 1 |
| 592 | 119.11 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 16 |
| 997 | 119.89 | temperature | 20 | 720 | > th | dose dependent | sin | 10 | NA |