Os11g0496500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to AT.I.24-5 protein (Fragment).


FiT-DB / Search/ Help/ Sample detail
|
Os11g0496500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to AT.I.24-5 protein (Fragment).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
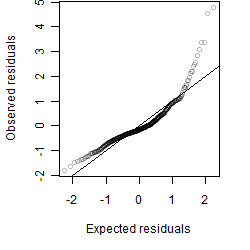
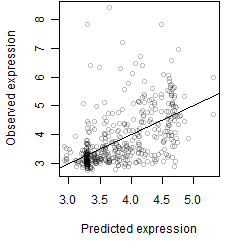 __
__

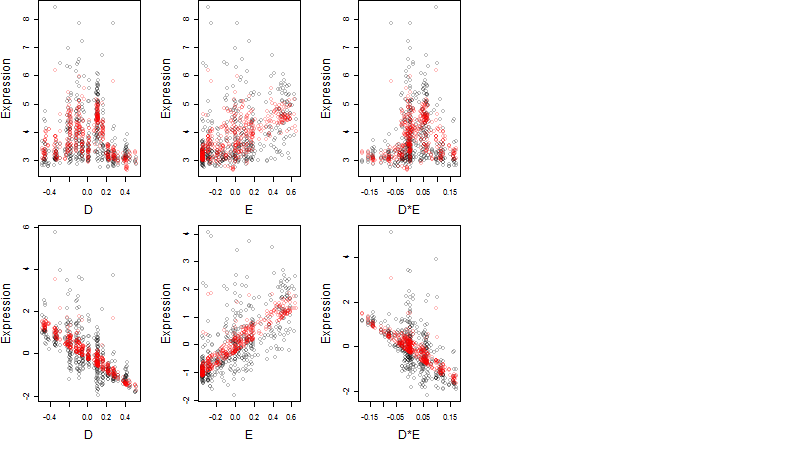
Dependence on each variable
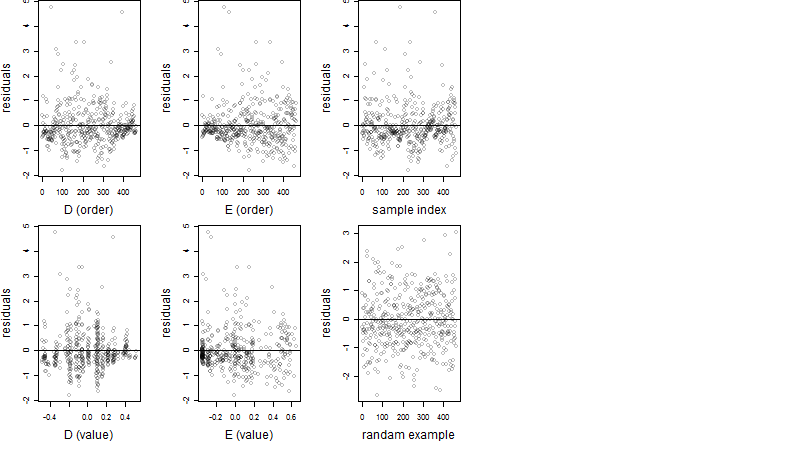
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 277.73 | 0.783 | 4.15 | -3 | 0.291 | 2.46 | -1.07 | -8.6 | -0.864 | 17.9 | 24.56 | 1263 | -- | -- |
| 303.17 | 0.811 | 3.92 | -0.956 | 0.287 | 2.4 | -1.12 | -- | -0.482 | 18.6 | 5.708 | 1231 | -- | -- |
| 280.35 | 0.78 | 4.15 | -2.95 | 0.293 | 2.45 | -- | -8.28 | -0.899 | 17.9 | 24.21 | 1243 | -- | -- |
| 306.13 | 0.815 | 3.92 | -0.912 | 0.307 | 2.32 | -- | -- | -0.486 | 18.5 | 15.81 | 1244 | -- | -- |
| 284.4 | 0.785 | 4.15 | -2.95 | -- | 2.44 | -- | -8.36 | -0.914 | -- | 24.16 | 1216 | -- | -- |
| 397.8 | 0.935 | 3.83 | -0.236 | 0.347 | -- | -0.589 | -- | -0.0567 | 21.2 | -- | -- | -- | -- |
| 398.73 | 0.935 | 3.83 | -0.218 | 0.35 | -- | -- | -- | -0.0581 | 22 | -- | -- | -- | -- |
| 306.18 | 0.815 | 3.9 | -0.791 | -- | 1.83 | -- | -- | -0.437 | -- | 20.99 | 1035 | -- | -- |
| 319.68 | 0.833 | 3.88 | -- | 0.197 | 1.53 | -- | -- | -0.32 | 19.2 | 21.62 | 1144 | -- | -- |
| 406.07 | 0.942 | 3.82 | -0.214 | -- | -- | -- | -- | -0.058 | -- | -- | -- | -- | -- |
| 399.98 | 0.936 | 3.82 | -- | 0.349 | -- | -- | -- | -0.0338 | 21.9 | -- | -- | -- | -- |
| 321.04 | 0.835 | 3.87 | -- | -- | 1.62 | -- | -- | -0.301 | -- | 21.62 | 1009 | -- | -- |
| 407.28 | 0.942 | 3.81 | -- | -- | -- | -- | -- | -0.0342 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance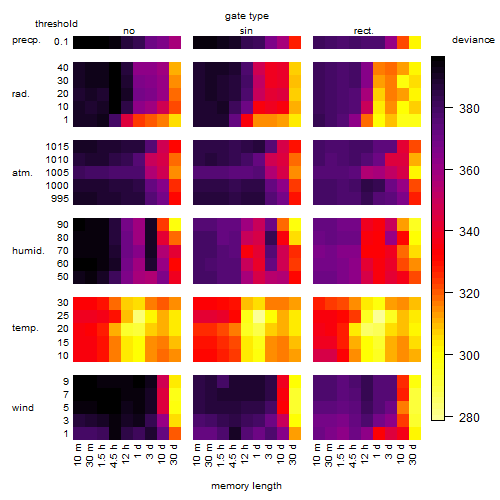
|
Histogram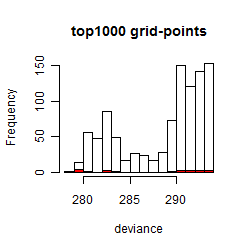 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 278.55 | temperature | 25 | 1440 | > th | dose dependent | rect. | 16 | 1 |
| 2 | 279.08 | temperature | 25 | 1440 | > th | dose dependent | rect. | 15 | 18 |
| 3 | 279.51 | temperature | 25 | 1440 | > th | dose dependent | rect. | 16 | 3 |
| 5 | 279.57 | temperature | 25 | 1440 | > th | dose dependent | rect. | 15 | 9 |
| 12 | 279.92 | temperature | 25 | 1440 | > th | dose dependent | sin | 16 | NA |
| 42 | 280.62 | temperature | 25 | 1440 | > th | dose dependent | rect. | 12 | 12 |
| 123 | 282.28 | temperature | 25 | 1440 | > th | dose dependent | rect. | 7 | 17 |
| 180 | 282.63 | temperature | 25 | 1440 | > th | dose dependent | no | NA | NA |
| 187 | 282.71 | temperature | 25 | 1440 | > th | dose dependent | rect. | 21 | 23 |
| 235 | 283.52 | temperature | 25 | 1440 | > th | dose dependent | rect. | 18 | 23 |
| 410 | 289.74 | wind | 5 | 43200 | > th | dose dependent | rect. | 9 | 7 |
| 485 | 290.38 | wind | 7 | 43200 | < th | dose independent | rect. | 5 | 11 |
| 491 | 290.46 | wind | 7 | 43200 | > th | dose independent | rect. | 5 | 11 |
| 624 | 291.30 | wind | 7 | 43200 | > th | dose independent | rect. | 24 | 16 |
| 627 | 291.32 | wind | 5 | 43200 | > th | dose dependent | rect. | 5 | 12 |
| 681 | 291.72 | wind | 5 | 43200 | > th | dose dependent | sin | 23 | NA |
| 727 | 292.16 | wind | 7 | 43200 | < th | dose independent | sin | 1 | NA |
| 732 | 292.19 | wind | 7 | 43200 | > th | dose independent | sin | 2 | NA |
| 845 | 292.99 | wind | 5 | 43200 | > th | dose independent | rect. | 10 | 6 |
| 922 | 293.42 | humidity | 90 | 43200 | > th | dose independent | rect. | 22 | 1 |
| 977 | 293.71 | wind | 5 | 43200 | < th | dose independent | rect. | 10 | 5 |
| 985 | 293.77 | wind | 7 | 43200 | > th | dose independent | rect. | 20 | 21 |