Os11g0153400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Rac GTPase activating protein 3 (Fragment).
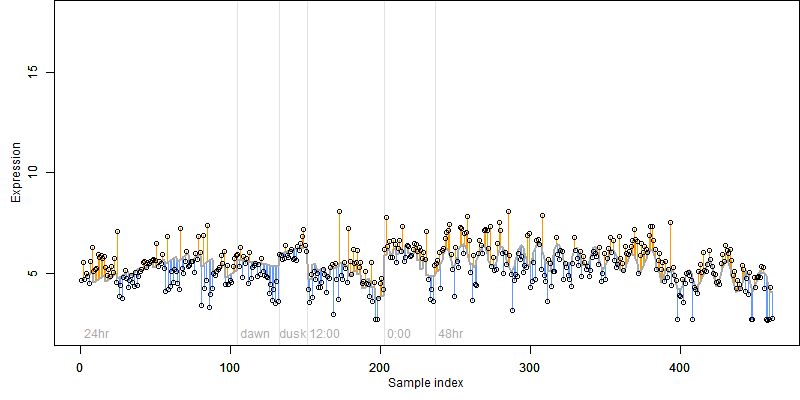

FiT-DB / Search/ Help/ Sample detail
|
Os11g0153400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Rac GTPase activating protein 3 (Fragment).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
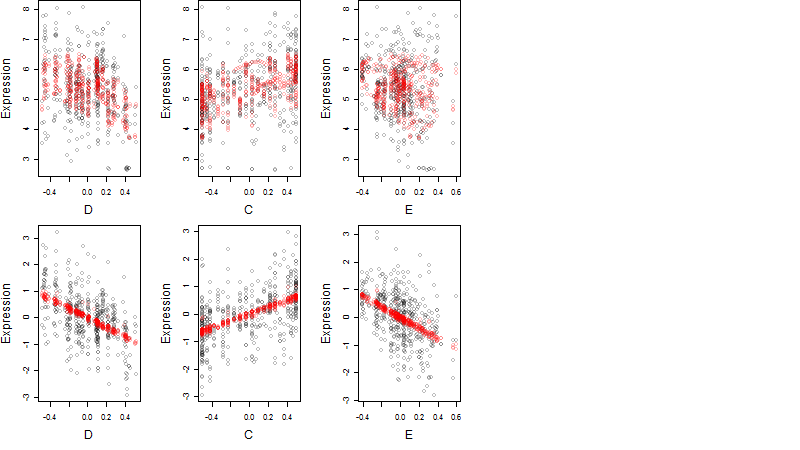
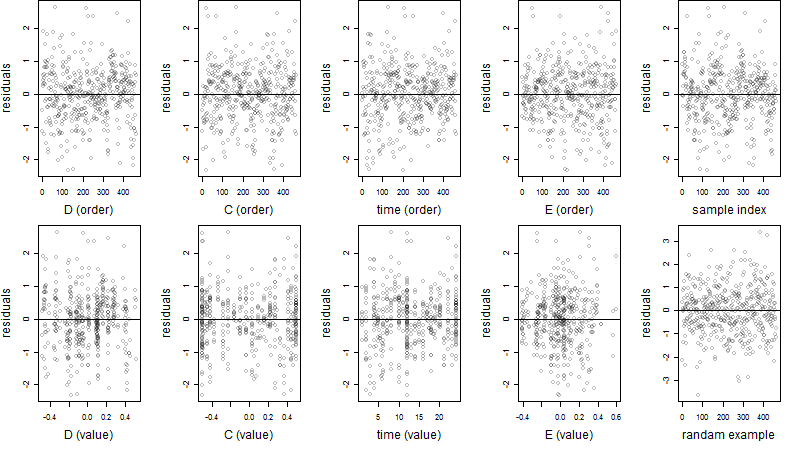
Dependence on each variable
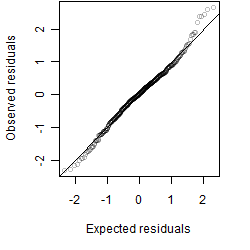
Residual plot
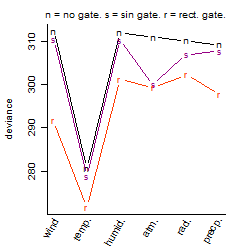
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 280.34 | 0.787 | 5.46 | -1.11 | 1.23 | -1.83 | -0.834 | -3.96 | -0.516 | 0.444 | 30.79 | 242 | -- | -- |
| 294.45 | 0.799 | 5.55 | -1.79 | 1.23 | -1.96 | -1.39 | -- | -0.737 | 0.648 | 31.96 | 244 | -- | -- |
| 281.3 | 0.781 | 5.45 | -1 | 1.33 | -1.79 | -- | -4.42 | -0.478 | 0.135 | 30.75 | 154 | -- | -- |
| 302.9 | 0.811 | 5.54 | -1.76 | 1.25 | -1.91 | -- | -- | -0.67 | 0.243 | 32.2 | 216 | -- | -- |
| 334.91 | 0.852 | 5.45 | -1.23 | -- | -2.36 | -- | -3.62 | -0.59 | -- | 31.36 | 727 | -- | -- |
| 341.57 | 0.866 | 5.47 | -1.34 | 1.05 | -- | -0.908 | -- | -0.359 | 22.8 | -- | -- | -- | -- |
| 344.37 | 0.869 | 5.47 | -1.3 | 1.07 | -- | -- | -- | -0.353 | 22.4 | -- | -- | -- | -- |
| 336.35 | 0.854 | 5.52 | -1.85 | -- | -2.22 | -- | -- | -0.69 | -- | 45.03 | 757 | -- | -- |
| 373.64 | 0.9 | 5.46 | -- | 1.15 | -1.18 | -- | -- | -0.344 | 23.4 | 40.28 | 160 | -- | -- |
| 413.82 | 0.951 | 5.44 | -1.29 | -- | -- | -- | -- | -0.35 | -- | -- | -- | -- | -- |
| 389.09 | 0.923 | 5.44 | -- | 1.06 | -- | -- | -- | -0.208 | 22.3 | -- | -- | -- | -- |
| 417.79 | 0.952 | 5.45 | -- | -- | -1.51 | -- | -- | -0.394 | -- | 45.56 | 727 | -- | -- |
| 457.39 | 0.998 | 5.41 | -- | -- | -- | -- | -- | -0.207 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance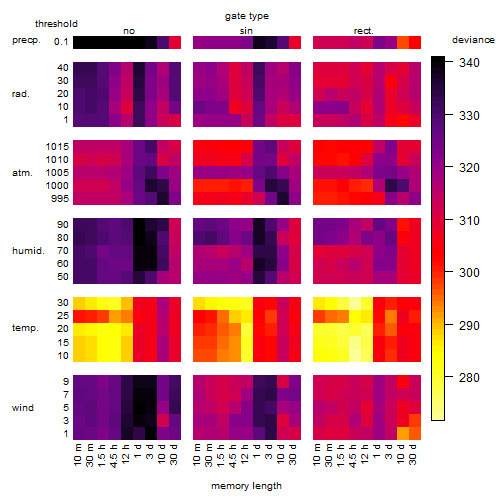
|
Histogram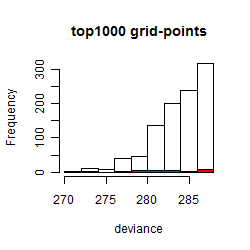 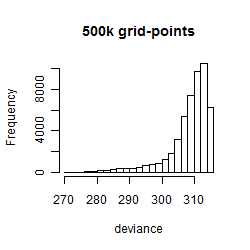
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 271.66 | temperature | 30 | 270 | < th | dose dependent | rect. | 22 | 21 |
| 4 | 272.49 | temperature | 10 | 270 | > th | dose dependent | rect. | 3 | 22 |
| 22 | 276.18 | temperature | 20 | 720 | > th | dose dependent | rect. | 22 | 19 |
| 27 | 276.80 | temperature | 10 | 720 | > th | dose dependent | rect. | 21 | 20 |
| 43 | 277.31 | temperature | 20 | 90 | > th | dose dependent | rect. | 6 | 22 |
| 69 | 278.67 | temperature | 30 | 270 | < th | dose dependent | rect. | 8 | 23 |
| 72 | 278.81 | temperature | 20 | 720 | > th | dose dependent | sin | 5 | NA |
| 83 | 279.35 | temperature | 20 | 270 | > th | dose dependent | rect. | 15 | 22 |
| 106 | 279.96 | temperature | 15 | 270 | > th | dose dependent | rect. | 5 | 17 |
| 120 | 280.18 | temperature | 10 | 720 | > th | dose dependent | sin | 4 | NA |
| 135 | 280.41 | temperature | 30 | 720 | < th | dose dependent | sin | 6 | NA |
| 162 | 280.72 | temperature | 30 | 270 | < th | dose dependent | no | NA | NA |
| 224 | 281.65 | temperature | 20 | 30 | > th | dose dependent | rect. | 21 | 23 |
| 284 | 282.51 | temperature | 15 | 30 | > th | dose dependent | rect. | 3 | 23 |
| 299 | 282.75 | temperature | 20 | 90 | > th | dose dependent | no | NA | NA |
| 300 | 282.76 | temperature | 30 | 90 | < th | dose dependent | sin | 5 | NA |
| 360 | 283.36 | temperature | 10 | 270 | > th | dose dependent | no | NA | NA |
| 382 | 283.55 | temperature | 20 | 30 | > th | dose dependent | rect. | 23 | 23 |
| 663 | 285.88 | temperature | 25 | 720 | > th | dose independent | sin | 3 | NA |
| 666 | 285.92 | temperature | 25 | 720 | > th | dose independent | rect. | 24 | 19 |
| 733 | 286.36 | temperature | 15 | 720 | > th | dose dependent | rect. | 5 | 6 |
| 766 | 286.57 | temperature | 25 | 720 | < th | dose independent | sin | 5 | NA |
| 823 | 286.91 | temperature | 20 | 30 | > th | dose dependent | rect. | 23 | 21 |
| 850 | 287.08 | temperature | 25 | 720 | < th | dose independent | rect. | 18 | 19 |
| 886 | 287.31 | temperature | 25 | 720 | < th | dose independent | rect. | 1 | 12 |
| 897 | 287.36 | temperature | 25 | 720 | > th | dose independent | rect. | 2 | 13 |
| 950 | 287.62 | temperature | 30 | 30 | < th | dose dependent | rect. | 1 | 23 |
| 965 | 287.69 | temperature | 25 | 720 | < th | dose independent | rect. | 24 | 15 |