Os11g0150400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Dimeric alpha-beta barrel domain containing protein.

FiT-DB / Search/ Help/ Sample detail
|
Os11g0150400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Dimeric alpha-beta barrel domain containing protein.
|
|
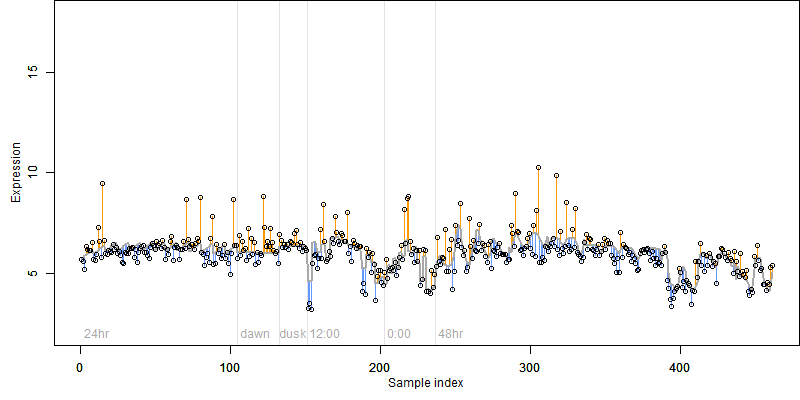
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
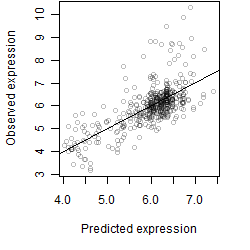 __
__

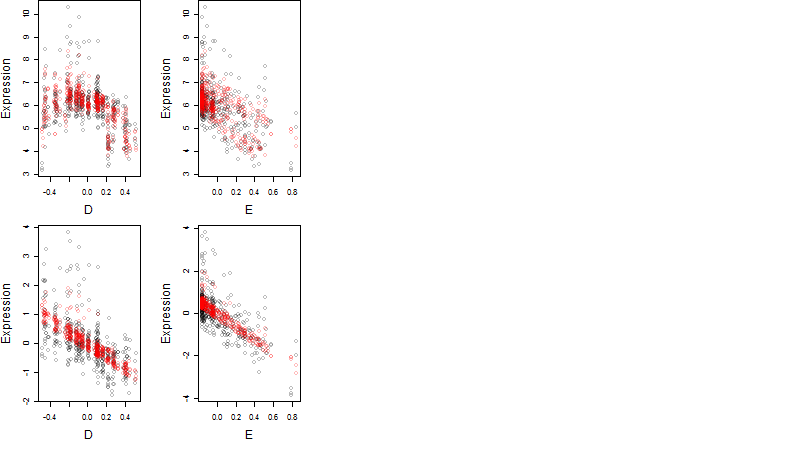
Dependence on each variable
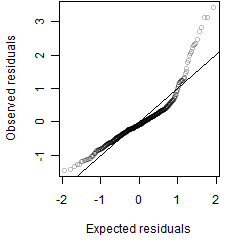
Residual plot
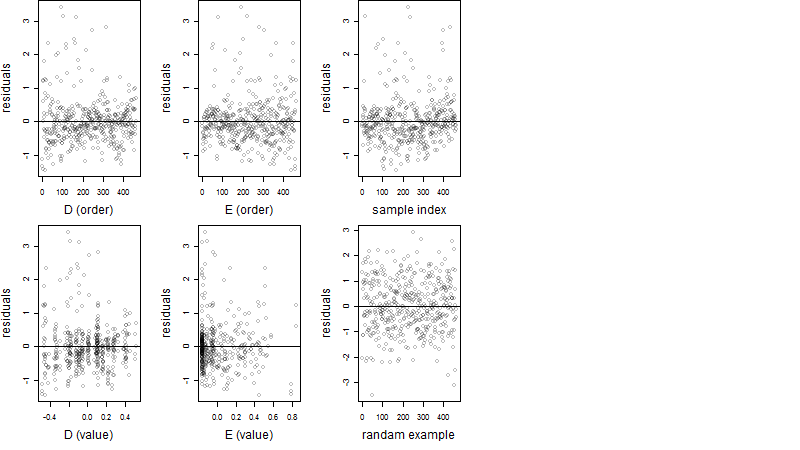
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 206.01 | 0.674 | 6.09 | -2.15 | 0.276 | -3.13 | -0.902 | 0.661 | -0.496 | 1.41 | 25.74 | 203 | -- | -- |
| 206.56 | 0.669 | 6.08 | -2.04 | 0.281 | -3.18 | -0.753 | -- | -0.471 | 1.28 | 25.7 | 200 | -- | -- |
| 208.85 | 0.673 | 6.09 | -2.13 | 0.336 | -3.18 | -- | 0.552 | -0.505 | 2.25 | 25.98 | 200 | -- | -- |
| 208.55 | 0.673 | 6.08 | -2.02 | 0.32 | -3.21 | -- | -- | -0.483 | 2.39 | 25.9 | 203 | -- | -- |
| 212.27 | 0.679 | 6.07 | -2.05 | -- | -3.02 | -- | 0.444 | -0.443 | -- | 25.6 | 249 | -- | -- |
| 358.38 | 0.887 | 5.92 | -1.21 | 0.336 | -- | 0.547 | -- | 0.209 | 15.8 | -- | -- | -- | -- |
| 358.89 | 0.887 | 5.92 | -1.2 | 0.356 | -- | -- | -- | 0.212 | 16.7 | -- | -- | -- | -- |
| 212.23 | 0.678 | 6.07 | -1.98 | -- | -3.07 | -- | -- | -0.433 | -- | 25.5 | 244 | -- | -- |
| 304.89 | 0.813 | 6 | -- | 0.189 | -2.42 | -- | -- | -0.123 | 24 | 25.8 | 159 | -- | -- |
| 365.6 | 0.893 | 5.92 | -1.21 | -- | -- | -- | -- | 0.203 | -- | -- | -- | -- | -- |
| 396.83 | 0.932 | 5.89 | -- | 0.378 | -- | -- | -- | 0.346 | 16.6 | -- | -- | -- | -- |
| 306 | 0.815 | 5.99 | -- | -- | -2.32 | -- | -- | -0.0832 | -- | 25.7 | 200 | -- | -- |
| 404.45 | 0.939 | 5.89 | -- | -- | -- | -- | -- | 0.337 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance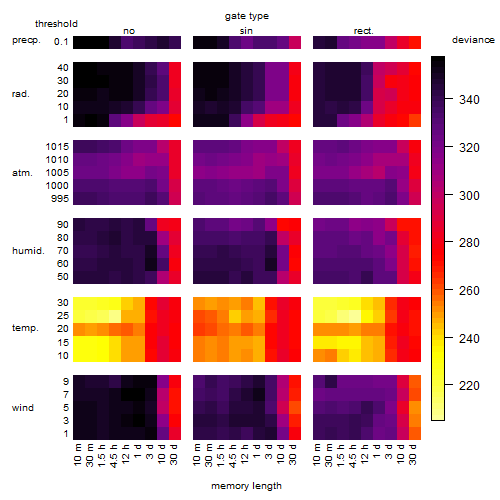
|
Histogram 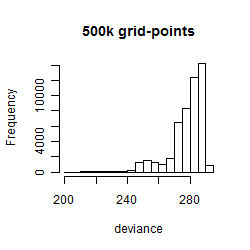
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 204.99 | temperature | 25 | 270 | < th | dose dependent | rect. | 6 | 23 |
| 10 | 209.71 | temperature | 25 | 270 | < th | dose dependent | no | NA | NA |
| 13 | 210.50 | temperature | 25 | 270 | < th | dose dependent | rect. | 15 | 23 |
| 14 | 210.63 | temperature | 25 | 270 | < th | dose dependent | rect. | 17 | 23 |
| 16 | 210.91 | temperature | 25 | 270 | < th | dose dependent | rect. | 20 | 23 |
| 18 | 210.93 | temperature | 25 | 270 | < th | dose dependent | rect. | 13 | 23 |
| 39 | 214.49 | temperature | 25 | 270 | < th | dose dependent | rect. | 24 | 23 |
| 42 | 214.51 | temperature | 25 | 270 | < th | dose dependent | rect. | 10 | 23 |
| 75 | 218.61 | temperature | 25 | 30 | < th | dose dependent | rect. | 1 | 23 |
| 77 | 218.81 | temperature | 25 | 30 | < th | dose dependent | rect. | 23 | 23 |
| 82 | 218.93 | temperature | 25 | 30 | < th | dose dependent | rect. | 11 | 23 |
| 310 | 230.14 | temperature | 15 | 10 | > th | dose dependent | no | NA | NA |
| 437 | 234.95 | temperature | 15 | 90 | > th | dose dependent | rect. | 2 | 23 |
| 440 | 235.08 | temperature | 15 | 270 | > th | dose dependent | rect. | 13 | 23 |
| 472 | 236.13 | temperature | 15 | 10 | > th | dose dependent | rect. | 2 | 23 |
| 676 | 242.25 | temperature | 25 | 270 | < th | dose dependent | sin | 21 | NA |
| 690 | 242.56 | temperature | 15 | 270 | > th | dose dependent | rect. | 11 | 23 |
| 700 | 242.75 | temperature | 25 | 1440 | < th | dose dependent | rect. | 4 | 8 |
| 848 | 245.44 | temperature | 30 | 1440 | < th | dose dependent | sin | 4 | NA |
| 874 | 245.66 | temperature | 30 | 1440 | < th | dose dependent | rect. | 21 | 17 |
| 899 | 245.81 | temperature | 30 | 1440 | < th | dose dependent | rect. | 21 | 15 |
| 906 | 245.84 | temperature | 10 | 1440 | > th | dose dependent | rect. | 3 | 9 |
| 995 | 246.24 | temperature | 30 | 1440 | < th | dose dependent | rect. | 21 | 21 |