Os10g0557900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Peptidase M10A and M12B, matrixin and adamalysin family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os10g0557900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Peptidase M10A and M12B, matrixin and adamalysin family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
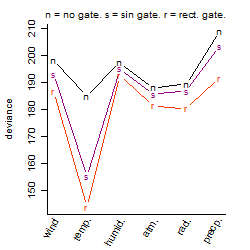
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 153.96 | 0.583 | 9.95 | -0.199 | 1.41 | 1.67 | -0.333 | -1.02 | 0.0862 | 1.66 | 21.11 | 706 | 0.302 | -- |
| 155.42 | 0.581 | 9.93 | -0.00329 | 1.45 | 1.7 | -0.0479 | -- | 0.14 | 1.67 | 21.1 | 711 | 0.248 | -- |
| 154.24 | 0.578 | 9.95 | -0.166 | 1.46 | 1.72 | -- | -0.823 | 0.0832 | 1.74 | 21.1 | 704 | 0.0998 | -- |
| 155.15 | 0.58 | 9.92 | -0.00318 | 1.5 | 1.74 | -- | -- | 0.134 | 1.84 | 21.15 | 707 | 0.00216 | -- |
| 202.62 | 0.663 | 9.99 | 0.00221 | -- | 1.12 | -- | -0.387 | -0.0321 | -- | 21.6 | 1877 | 3.04 | -- |
| 220.16 | 0.696 | 9.83 | 0.451 | 0.722 | -- | -0.53 | -- | 0.481 | 3.64 | -- | -- | -- | -- |
| 221 | 0.696 | 9.83 | 0.46 | 0.72 | -- | -- | -- | 0.484 | 3.7 | -- | -- | -- | -- |
| 195.33 | 0.651 | 9.92 | -0.0107 | -- | 1.25 | -- | -- | 0.116 | -- | 21.46 | 2286 | 7.24 | -- |
| 155.14 | 0.58 | 9.92 | -- | 1.5 | 1.73 | -- | -- | 0.134 | 1.84 | 21.15 | 707 | 2.18e-06 | -- |
| 249.74 | 0.738 | 9.83 | 0.49 | -- | -- | -- | -- | 0.503 | -- | -- | -- | -- | -- |
| 226.57 | 0.704 | 9.84 | -- | 0.729 | -- | -- | -- | 0.433 | 3.69 | -- | -- | -- | -- |
| 195.32 | 0.651 | 9.92 | -- | -- | 1.23 | -- | -- | 0.112 | -- | 21.45 | 2272 | 7.07 | -- |
| 256.07 | 0.747 | 9.84 | -- | -- | -- | -- | -- | 0.449 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 143.69 | temperature | 15 | 720 | > th | dose dependent | rect. | 1 | 6 |
| 5 | 144.67 | temperature | 30 | 720 | < th | dose dependent | rect. | 24 | 8 |
| 20 | 146.20 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 4 |
| 32 | 147.22 | temperature | 20 | 720 | > th | dose independent | rect. | 3 | 4 |
| 54 | 149.15 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 2 |
| 122 | 152.94 | temperature | 20 | 720 | < th | dose independent | rect. | 3 | 4 |
| 156 | 154.48 | temperature | 20 | 720 | < th | dose independent | rect. | 5 | 2 |
| 173 | 155.06 | temperature | 25 | 720 | < th | dose dependent | sin | 7 | NA |
| 281 | 159.20 | temperature | 15 | 720 | > th | dose dependent | sin | 7 | NA |
| 418 | 163.57 | temperature | 20 | 720 | > th | dose independent | sin | 6 | NA |
| 442 | 164.34 | temperature | 20 | 720 | < th | dose independent | sin | 6 | NA |
| 509 | 165.99 | temperature | 20 | 720 | > th | dose independent | rect. | 23 | 14 |
| 715 | 170.73 | temperature | 25 | 270 | < th | dose independent | rect. | 7 | 5 |
| 958 | 175.33 | temperature | 10 | 270 | > th | dose dependent | rect. | 3 | 15 |