Os10g0409400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Polygalacturonase isoenzyme 1 beta subunit precursor.


FiT-DB / Search/ Help/ Sample detail
|
Os10g0409400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Polygalacturonase isoenzyme 1 beta subunit precursor.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
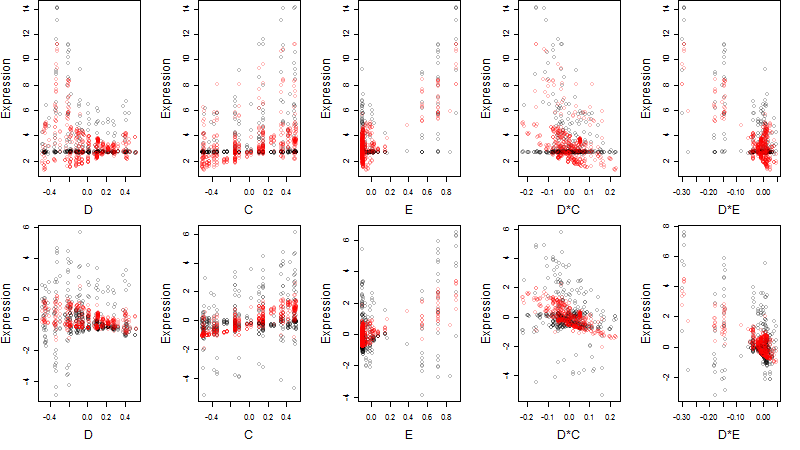
Dependence on each variable
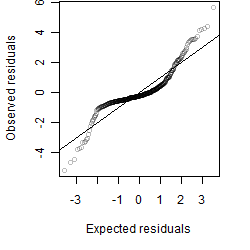
Residual plot

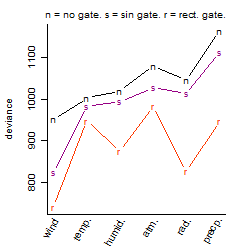
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose independent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 693.41 | 1.24 | 3.18 | -0.77 | 0.998 | 2.36 | -4.38 | -11 | -0.0648 | 4.85 | 7.194 | 2831 | 17.1 | 2.4 |
| 725.73 | 1.25 | 3.35 | -0.0977 | 0.929 | 5.06 | -4.65 | -- | -0.093 | 4.96 | 7 | 2640 | 16.9 | 1.85 |
| 742.01 | 1.27 | 3.17 | -0.734 | 0.984 | 2.31 | -- | -11.1 | -0.0359 | 4.79 | 7.2 | 2811 | 17.1 | 2.49 |
| 775.03 | 1.3 | 3.38 | -0.119 | 0.897 | 5.3 | -- | -- | -0.0687 | 4.86 | 7.1 | 2643 | 17.1 | 1.87 |
| 790.34 | 1.31 | 3.15 | -0.716 | -- | 2.13 | -- | -12.2 | 0.0206 | -- | 7.2 | 2829 | 17.1 | 2.48 |
| 1206.71 | 1.63 | 3.57 | -2.09 | 1.22 | -- | -5.7 | -- | -0.84 | 4.2 | -- | -- | -- | -- |
| 1297.58 | 1.69 | 3.55 | -2.03 | 1.18 | -- | -- | -- | -0.808 | 4.1 | -- | -- | -- | -- |
| 819.37 | 1.33 | 3.39 | 0.0129 | -- | 5.51 | -- | -- | -0.00378 | -- | 7.09 | 2641 | 17.1 | 1.8 |
| 774.88 | 1.3 | 3.38 | -- | 0.919 | 5.4 | -- | -- | -0.0308 | 4.81 | 7.089 | 2620 | 17.1 | 1.87 |
| 1373.79 | 1.73 | 3.56 | -1.99 | -- | -- | -- | -- | -0.777 | -- | -- | -- | -- | -- |
| 1406.68 | 1.75 | 3.5 | -- | 1.15 | -- | -- | -- | -0.582 | 4.13 | -- | -- | -- | -- |
| 813.06 | 1.33 | 3.38 | -- | -- | 5.68 | -- | -- | -0.0011 | -- | 7.07 | 2637 | 17.1 | 1.56 |
| 1477.89 | 1.79 | 3.51 | -- | -- | -- | -- | -- | -0.556 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance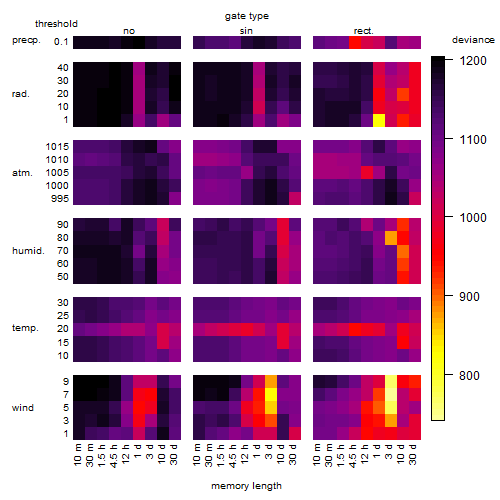
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 741.38 | wind | 7 | 4320 | > th | dose independent | rect. | 17 | 3 |
| 4 | 749.52 | wind | 7 | 4320 | < th | dose independent | rect. | 17 | 4 |
| 6 | 754.72 | wind | 7 | 4320 | > th | dose dependent | rect. | 17 | 2 |
| 9 | 758.61 | wind | 7 | 4320 | < th | dose independent | rect. | 18 | 1 |
| 42 | 788.59 | wind | 7 | 4320 | > th | dose independent | rect. | 15 | 11 |
| 61 | 793.93 | wind | 7 | 4320 | < th | dose independent | rect. | 15 | 11 |
| 183 | 824.26 | wind | 7 | 4320 | > th | dose independent | sin | 14 | NA |
| 209 | 827.47 | radiation | 1 | 1440 | < th | dose independent | rect. | 11 | 1 |
| 210 | 827.47 | radiation | 1 | 1440 | < th | dose dependent | rect. | 11 | 1 |
| 217 | 828.07 | wind | 7 | 4320 | < th | dose independent | sin | 14 | NA |
| 272 | 837.30 | wind | 5 | 4320 | < th | dose independent | rect. | 21 | 1 |
| 319 | 844.13 | wind | 5 | 4320 | < th | dose independent | rect. | 24 | 1 |
| 344 | 850.98 | wind | 5 | 4320 | > th | dose independent | rect. | 21 | 1 |
| 349 | 851.04 | wind | 5 | 4320 | > th | dose independent | rect. | 24 | 1 |
| 361 | 852.56 | wind | 5 | 4320 | > th | dose dependent | sin | 14 | NA |
| 448 | 863.99 | radiation | 1 | 1440 | < th | dose independent | rect. | 13 | 2 |
| 449 | 863.99 | radiation | 1 | 1440 | < th | dose dependent | rect. | 13 | 2 |
| 457 | 864.99 | radiation | 1 | 1440 | < th | dose dependent | rect. | 11 | 5 |
| 475 | 867.50 | radiation | 1 | 1440 | > th | dose independent | rect. | 13 | 2 |
| 533 | 875.40 | humidity | 80 | 4320 | < th | dose independent | rect. | 22 | 1 |
| 553 | 877.74 | wind | 3 | 4320 | > th | dose dependent | rect. | 24 | 1 |
| 641 | 887.50 | humidity | 80 | 4320 | > th | dose independent | rect. | 22 | 1 |
| 697 | 895.82 | humidity | 70 | 14400 | < th | dose dependent | rect. | 21 | 1 |
| 906 | 916.94 | humidity | 60 | 14400 | < th | dose independent | rect. | 6 | 1 |
| 908 | 916.94 | humidity | 60 | 14400 | < th | dose independent | rect. | 20 | 1 |
| 910 | 916.94 | radiation | 20 | 14400 | > th | dose independent | rect. | 17 | 1 |
| 961 | 916.94 | humidity | 60 | 14400 | < th | dose dependent | rect. | 6 | 1 |
| 965 | 916.94 | radiation | 20 | 14400 | > th | dose dependent | rect. | 17 | 1 |