FiT-DB /
Search/
Help/
Sample detail
|
Os09g0505700
|
blastp(Os)/
blastp(At)/
coex///
RAP/
RiceXPro/
SALAD/
ATTED-II
|
|
Description : Ribulose-phosphate 3-epimerase, cytoplasmic isoform (EC 5.1.3.1) (Ribulose-5-phosphate-epimerase) (Cyt-RPEase) (RPEcyt) (Pentose-5- phosphate 3-epimerase) (PPE).


|
log2(Expression) ~
Norm(μ, σ2)
μ = α
+ β1D
+ β2C
+ β3E
+ β4D*C
+ β5D*E
+ γ1N8
| par. |
|
value |
(S.E.) |
|
|
|
|
|
|
| α |
: |
12.7 |
(0.0187) |
R2 |
: |
0.0625 |
|
|
|
| β1 |
: |
-- |
(--) |
R2dD |
: |
-- |
R2D |
: |
-- |
| β2 |
: |
-- |
(--) |
R2dC |
: |
-- |
R2C |
: |
-- |
| β3 |
: |
-- |
(--) |
R2dE |
: |
-- |
R2E |
: |
-- |
| β4 |
: |
-- |
(--) |
R2dD*C |
: |
-- |
R2D*C |
: |
-- |
| β5 |
: |
-- |
(--) |
R2dD*E |
: |
-- |
R2D*E |
: |
-- |
| γ1 |
: |
0.218 |
(0.0395) |
R2dN8 |
: |
0.0808 |
R2N8 |
: |
0.0625 |
| σ |
: |
0.353 |
|
|
|
|
|
|
|
| deviance |
: |
57.23 |
|
|
|
|
|
|
|
| __ |
|
|
|
|
|
|
|
|
|
|
_____
|
| C |
|
|
| peak time of C |
: |
-- |
| E |
|
|
| wheather |
: |
-- |
| threshold |
: |
-- |
| memory length |
: |
-- |
| response mode |
: |
-- |
| dose dependency |
: |
-- |
| G |
|
|
| type of G |
: |
-- |
| peak or start time of G |
: |
-- |
| open length of G |
: |
-- |
|

 __
__
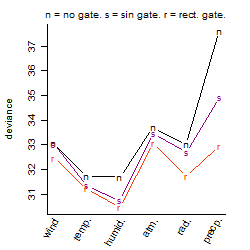
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
| 31.47 |
0.264 |
12.8 |
-0.838 |
0.434 |
-0.679 |
-0.0475 |
1 |
-0.00785 |
23.4 |
23.49 |
1531 |
-- |
-- |
| 32.26 |
0.265 |
12.8 |
-0.709 |
0.432 |
-0.898 |
-0.0466 |
-- |
0.0257 |
23.4 |
22.56 |
1474 |
-- |
-- |
| 31.47 |
0.261 |
12.8 |
-0.833 |
0.434 |
-0.687 |
-- |
0.991 |
-0.00657 |
23.5 |
23.33 |
1530 |
-- |
-- |
| 32.27 |
0.265 |
12.8 |
-0.701 |
0.426 |
-0.897 |
-- |
-- |
0.029 |
23.4 |
22.56 |
1440 |
-- |
-- |
| 37.94 |
0.287 |
12.8 |
-0.939 |
-- |
-0.849 |
-- |
1.31 |
-0.061 |
-- |
30.19 |
904 |
-- |
-- |
| 40.2 |
0.297 |
12.8 |
-0.452 |
0.436 |
-- |
-0.021 |
-- |
0.164 |
23.6 |
-- |
-- |
-- |
-- |
| 40.2 |
0.297 |
12.8 |
-0.452 |
0.436 |
-- |
-- |
-- |
0.164 |
23.6 |
-- |
-- |
-- |
-- |
| 39.4 |
0.292 |
12.8 |
-0.694 |
-- |
-0.887 |
-- |
-- |
-0.0164 |
-- |
29.1 |
888 |
-- |
-- |
| 42.78 |
0.306 |
12.8 |
-- |
0.495 |
-0.451 |
-- |
-- |
0.164 |
23.7 |
23.2 |
1314 |
-- |
-- |
| 52.13 |
0.337 |
12.8 |
-0.44 |
-- |
-- |
-- |
-- |
0.169 |
-- |
-- |
-- |
-- |
-- |
| 45.58 |
0.316 |
12.8 |
-- |
0.431 |
-- |
-- |
-- |
0.214 |
23.5 |
-- |
-- |
-- |
-- |
| 50.57 |
0.331 |
12.8 |
-- |
-- |
-0.601 |
-- |
-- |
0.114 |
-- |
29.5 |
859 |
-- |
-- |
| 57.23 |
0.353 |
12.7 |
-- |
-- |
-- |
-- |
-- |
0.218 |
-- |
-- |
-- |
-- |
-- |
Results of the grid search
Summarized heatmap of deviance

|
Histogram


|
Local optima within top1000 grid-points
| rank |
deviance |
wheather |
threshold |
memory length |
response mode |
dose dependency |
type of G |
peak or start time of G |
open length of G |
| 1 |
30.48 |
humidity |
90 |
14400 |
> th |
dose dependent |
rect. |
22 |
4 |
| 6 |
30.65 |
humidity |
90 |
14400 |
> th |
dose dependent |
rect. |
22 |
11 |
| 17 |
30.75 |
humidity |
90 |
14400 |
> th |
dose dependent |
sin |
9 |
NA |
| 84 |
30.94 |
humidity |
90 |
14400 |
> th |
dose dependent |
rect. |
4 |
3 |
| 121 |
31.04 |
humidity |
90 |
14400 |
> th |
dose dependent |
rect. |
3 |
5 |
| 177 |
31.22 |
temperature |
25 |
1440 |
< th |
dose dependent |
rect. |
21 |
1 |
| 193 |
31.28 |
temperature |
25 |
1440 |
< th |
dose dependent |
rect. |
18 |
16 |
| 207 |
31.31 |
temperature |
25 |
1440 |
< th |
dose dependent |
rect. |
18 |
12 |
| 232 |
31.35 |
temperature |
25 |
1440 |
< th |
dose dependent |
sin |
11 |
NA |
| 271 |
31.41 |
humidity |
90 |
14400 |
> th |
dose independent |
rect. |
21 |
10 |
| 301 |
31.44 |
temperature |
20 |
1440 |
< th |
dose dependent |
rect. |
2 |
3 |
| 325 |
31.47 |
temperature |
25 |
1440 |
< th |
dose dependent |
rect. |
2 |
8 |
| 350 |
31.50 |
temperature |
10 |
1440 |
> th |
dose dependent |
rect. |
21 |
1 |
| 376 |
31.52 |
temperature |
10 |
1440 |
> th |
dose dependent |
rect. |
19 |
11 |
| 382 |
31.53 |
temperature |
20 |
1440 |
< th |
dose independent |
rect. |
5 |
1 |
| 431 |
31.56 |
humidity |
90 |
14400 |
> th |
dose independent |
rect. |
21 |
7 |
| 497 |
31.58 |
temperature |
10 |
720 |
> th |
dose dependent |
rect. |
9 |
22 |
| 536 |
31.61 |
temperature |
10 |
1440 |
> th |
dose dependent |
sin |
11 |
NA |
| 578 |
31.63 |
temperature |
10 |
1440 |
> th |
dose dependent |
rect. |
2 |
4 |
| 618 |
31.65 |
temperature |
20 |
1440 |
> th |
dose independent |
rect. |
5 |
2 |
| 620 |
31.65 |
humidity |
90 |
14400 |
< th |
dose independent |
rect. |
21 |
10 |
| 734 |
31.70 |
humidity |
90 |
14400 |
> th |
dose dependent |
no |
NA |
NA |
| 828 |
31.74 |
radiation |
30 |
4320 |
< th |
dose independent |
rect. |
16 |
16 |
| 829 |
31.74 |
temperature |
25 |
1440 |
< th |
dose dependent |
no |
NA |
NA |
| 876 |
31.76 |
humidity |
90 |
14400 |
> th |
dose independent |
sin |
9 |
NA |
| 919 |
31.77 |
temperature |
25 |
1440 |
< th |
dose dependent |
rect. |
7 |
23 |
| 927 |
31.78 |
humidity |
50 |
14400 |
> th |
dose dependent |
rect. |
23 |
4 |
| 941 |
31.78 |
temperature |
25 |
1440 |
< th |
dose dependent |
rect. |
1 |
23 |
| 943 |
31.78 |
temperature |
25 |
1440 |
< th |
dose dependent |
rect. |
23 |
23 |
| 957 |
31.79 |
humidity |
50 |
14400 |
> th |
dose dependent |
rect. |
22 |
6 |



 __
__





