Os09g0347500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Protein transport protein Sec61 alpha subunit isoform 2 (Sec61 alpha- 2).
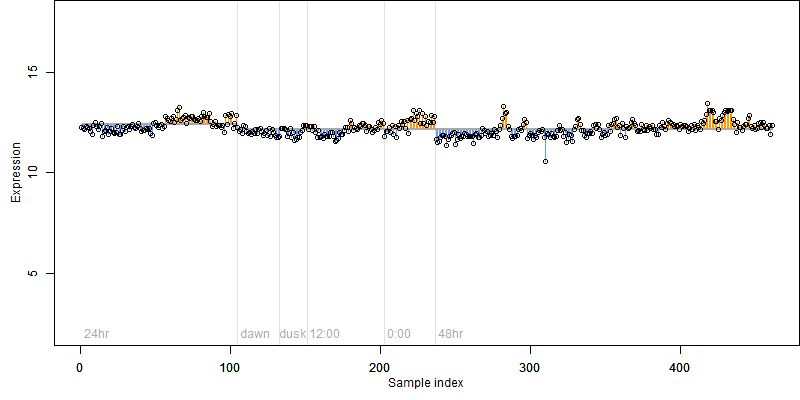

FiT-DB / Search/ Help/ Sample detail
|
Os09g0347500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Protein transport protein Sec61 alpha subunit isoform 2 (Sec61 alpha- 2).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
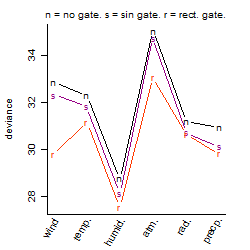
Dependence on each variable

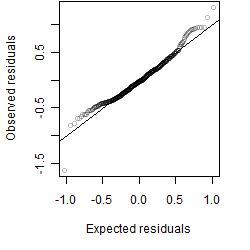
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 28.34 | 0.25 | 12.2 | 0.66 | 0.473 | 0.562 | 0.192 | 0.514 | 0.309 | 0.519 | 45.84 | 1302 | -- | -- |
| 28.74 | 0.25 | 12.2 | 0.711 | 0.481 | 0.599 | 0.161 | -- | 0.322 | 0.556 | 45.87 | 1292 | -- | -- |
| 28.49 | 0.249 | 12.2 | 0.658 | 0.471 | 0.562 | -- | 0.462 | 0.309 | 0.58 | 44.99 | 1305 | -- | -- |
| 28.81 | 0.25 | 12.2 | 0.711 | 0.477 | 0.615 | -- | -- | 0.319 | 0.587 | 39.91 | 1295 | -- | -- |
| 37.72 | 0.286 | 12.2 | 0.686 | -- | 0.559 | -- | 0.225 | 0.316 | -- | 39.33 | 1606 | -- | -- |
| 36.89 | 0.285 | 12.2 | 0.675 | 0.388 | -- | 0.0603 | -- | 0.296 | 0.252 | -- | -- | -- | -- |
| 36.91 | 0.284 | 12.2 | 0.672 | 0.388 | -- | -- | -- | 0.296 | 0.281 | -- | -- | -- | -- |
| 37.82 | 0.286 | 12.2 | 0.707 | -- | 0.591 | -- | -- | 0.325 | -- | 28.81 | 1619 | -- | -- |
| 41.26 | 0.299 | 12.2 | -- | 0.396 | 0.753 | -- | -- | 0.304 | 0.584 | 76.45 | 1441 | -- | -- |
| 46.38 | 0.318 | 12.2 | 0.685 | -- | -- | -- | -- | 0.302 | -- | -- | -- | -- | -- |
| 48.81 | 0.327 | 12.2 | -- | 0.398 | -- | -- | -- | 0.221 | 0.371 | -- | -- | -- | -- |
| 49.28 | 0.327 | 12.2 | -- | -- | 0.828 | -- | -- | 0.318 | -- | 73.99 | 1616 | -- | -- |
| 58.76 | 0.358 | 12.2 | -- | -- | -- | -- | -- | 0.226 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance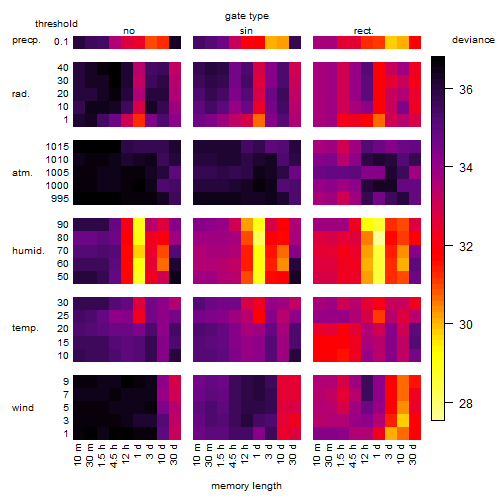
|
Histogram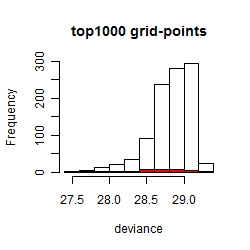 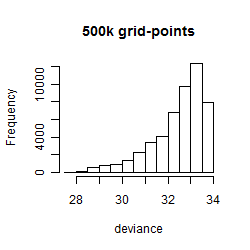
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 27.53 | humidity | 80 | 1440 | < th | dose independent | rect. | 11 | 2 |
| 2 | 27.63 | humidity | 80 | 1440 | > th | dose independent | rect. | 11 | 2 |
| 3 | 27.77 | humidity | 80 | 1440 | < th | dose independent | rect. | 7 | 13 |
| 5 | 27.78 | humidity | 80 | 1440 | < th | dose independent | rect. | 7 | 7 |
| 8 | 27.83 | humidity | 80 | 1440 | > th | dose independent | rect. | 7 | 14 |
| 29 | 28.13 | humidity | 80 | 1440 | < th | dose independent | sin | 1 | NA |
| 37 | 28.19 | humidity | 80 | 1440 | > th | dose independent | rect. | 7 | 7 |
| 56 | 28.31 | humidity | 80 | 1440 | > th | dose independent | sin | 0 | NA |
| 92 | 28.47 | humidity | 70 | 1440 | > th | dose dependent | rect. | 11 | 2 |
| 96 | 28.49 | humidity | 60 | 1440 | > th | dose dependent | rect. | 18 | 1 |
| 100 | 28.50 | humidity | 50 | 1440 | > th | dose dependent | rect. | 7 | 16 |
| 101 | 28.50 | humidity | 50 | 1440 | > th | dose dependent | rect. | 5 | 17 |
| 143 | 28.56 | humidity | 70 | 1440 | > th | dose dependent | rect. | 7 | 14 |
| 153 | 28.58 | humidity | 50 | 1440 | > th | dose dependent | rect. | 15 | 22 |
| 186 | 28.63 | humidity | 90 | 1440 | < th | dose dependent | rect. | 15 | 22 |
| 223 | 28.67 | humidity | 90 | 1440 | < th | dose dependent | rect. | 6 | 15 |
| 304 | 28.73 | humidity | 90 | 1440 | < th | dose dependent | rect. | 18 | 1 |
| 314 | 28.74 | humidity | 50 | 1440 | > th | dose dependent | no | NA | NA |
| 369 | 28.78 | humidity | 80 | 1440 | < th | dose independent | rect. | 17 | 21 |
| 395 | 28.80 | humidity | 80 | 1440 | > th | dose independent | rect. | 17 | 21 |
| 433 | 28.82 | humidity | 50 | 1440 | > th | dose dependent | rect. | 11 | 23 |
| 444 | 28.83 | humidity | 50 | 1440 | > th | dose dependent | rect. | 17 | 4 |
| 467 | 28.85 | humidity | 50 | 1440 | > th | dose dependent | sin | 22 | NA |
| 499 | 28.87 | humidity | 90 | 1440 | < th | dose dependent | no | NA | NA |
| 567 | 28.92 | humidity | 90 | 1440 | < th | dose dependent | rect. | 9 | 23 |
| 600 | 28.94 | humidity | 90 | 1440 | < th | dose dependent | rect. | 11 | 23 |
| 648 | 28.98 | humidity | 90 | 1440 | < th | dose dependent | sin | 21 | NA |
| 696 | 29.02 | humidity | 70 | 1440 | > th | dose dependent | rect. | 7 | 7 |
| 738 | 29.06 | humidity | 80 | 1440 | > th | dose independent | no | NA | NA |
| 773 | 29.09 | humidity | 80 | 1440 | < th | dose independent | no | NA | NA |
| 940 | 29.19 | humidity | 80 | 1440 | < th | dose dependent | sin | 16 | NA |
| 990 | 29.21 | humidity | 90 | 720 | > th | dose dependent | rect. | 6 | 17 |