Os07g0608800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Peroxisome assembly protein 10 (Peroxin-10) (AthPEX10) (Pex10p) (PER8).


FiT-DB / Search/ Help/ Sample detail
|
Os07g0608800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Peroxisome assembly protein 10 (Peroxin-10) (AthPEX10) (Pex10p) (PER8).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
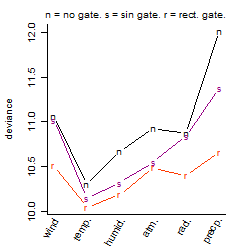
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10.2 | 0.15 | 11.2 | -0.773 | 0.389 | 0.191 | -0.139 | -0.55 | -0.0709 | 9.64 | 21.8 | 1857 | -- | -- |
| 10.88 | 0.154 | 11.2 | -0.603 | 0.399 | 0.201 | -0.11 | -- | -0.0432 | 9.61 | 22.2 | 2150 | -- | -- |
| 10.27 | 0.149 | 11.2 | -0.774 | 0.392 | 0.194 | -- | -0.546 | -0.0725 | 9.64 | 21.9 | 1859 | -- | -- |
| 10.84 | 0.153 | 11.2 | -0.608 | 0.39 | 0.232 | -- | -- | -0.0205 | 9.58 | 21.4 | 1377 | -- | -- |
| 18.35 | 0.201 | 11.2 | -0.607 | -- | 0.198 | -- | -0.128 | -0.0283 | -- | 22.01 | 1590 | -- | -- |
| 12.36 | 0.165 | 11.2 | -0.516 | 0.382 | -- | -0.11 | -- | 0.0349 | 9.64 | -- | -- | -- | -- |
| 12.41 | 0.165 | 11.2 | -0.52 | 0.382 | -- | -- | -- | 0.0347 | 9.62 | -- | -- | -- | -- |
| 18.46 | 0.201 | 11.2 | -0.574 | -- | 0.199 | -- | -- | -0.0203 | -- | 22.01 | 1590 | -- | -- |
| 19.34 | 0.206 | 11.2 | -- | 0.378 | 0.0517 | -- | -- | 0.0689 | 9.36 | 25.3 | 2392 | -- | -- |
| 21.05 | 0.214 | 11.2 | -0.523 | -- | -- | -- | -- | 0.0354 | -- | -- | -- | -- | -- |
| 19.53 | 0.207 | 11.2 | -- | 0.383 | -- | -- | -- | 0.0926 | 9.72 | -- | -- | -- | -- |
| 23.46 | 0.226 | 11.1 | -- | -- | -0.321 | -- | -- | 0.205 | -- | 22.06 | 47520 | -- | -- |
| 28.26 | 0.248 | 11.2 | -- | -- | -- | -- | -- | 0.0935 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance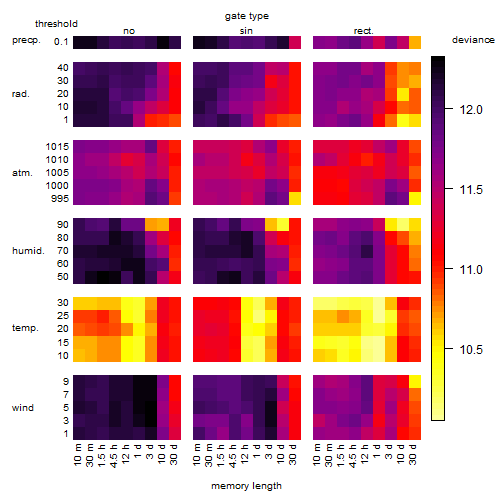
|
Histogram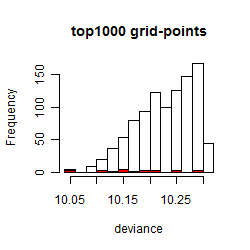 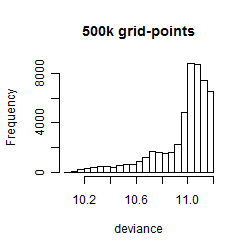
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10.05 | temperature | 10 | 1440 | > th | dose dependent | rect. | 5 | 1 |
| 2 | 10.05 | temperature | 25 | 1440 | < th | dose dependent | rect. | 5 | 1 |
| 21 | 10.11 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 11 |
| 33 | 10.12 | temperature | 15 | 720 | > th | dose dependent | rect. | 20 | 19 |
| 59 | 10.14 | temperature | 25 | 1440 | < th | dose dependent | rect. | 23 | 7 |
| 71 | 10.15 | temperature | 25 | 1440 | < th | dose dependent | sin | 9 | NA |
| 74 | 10.15 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 109 | 10.16 | temperature | 15 | 1440 | > th | dose dependent | rect. | 20 | 10 |
| 123 | 10.16 | temperature | 30 | 10 | < th | dose dependent | rect. | 22 | 22 |
| 155 | 10.17 | temperature | 15 | 720 | > th | dose dependent | rect. | 23 | 16 |
| 213 | 10.19 | humidity | 90 | 14400 | > th | dose dependent | rect. | 21 | 6 |
| 246 | 10.20 | temperature | 15 | 1440 | > th | dose dependent | sin | 10 | NA |
| 303 | 10.21 | temperature | 15 | 1440 | > th | dose dependent | rect. | 20 | 1 |
| 304 | 10.21 | humidity | 90 | 14400 | > th | dose dependent | rect. | 23 | 1 |
| 578 | 10.26 | temperature | 25 | 1440 | < th | dose dependent | rect. | 23 | 1 |
| 581 | 10.26 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 4 |
| 864 | 10.30 | temperature | 30 | 1440 | < th | dose dependent | no | NA | NA |
| 956 | 10.30 | temperature | 30 | 1440 | < th | dose dependent | rect. | 23 | 23 |