Os07g0604400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to BRITTLE CULM1.


FiT-DB / Search/ Help/ Sample detail
|
Os07g0604400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to BRITTLE CULM1.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

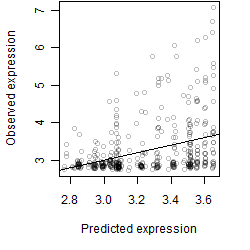 __
__
Dependence on each variable

Residual plot

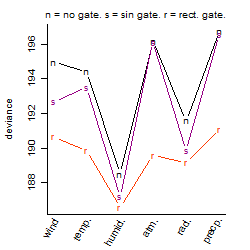
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 184.59 | 0.638 | 3.21 | -1.16 | 0.233 | -0.545 | -0.578 | 2.61 | 0.214 | 2.17 | 73.06 | 1681 | -- | -- |
| 195.94 | 0.652 | 3.17 | -0.84 | 0.18 | -0.403 | -0.157 | -- | 0.304 | 1.75 | 73.54 | 1669 | -- | -- |
| 185.63 | 0.635 | 3.21 | -1.14 | 0.239 | -0.537 | -- | 2.61 | 0.218 | 2.47 | 73 | 1742 | -- | -- |
| 196.01 | 0.652 | 3.17 | -0.824 | 0.184 | -0.414 | -- | -- | 0.311 | 2.47 | 73 | 1669 | -- | -- |
| 188.55 | 0.64 | 3.2 | -1.09 | -- | -0.443 | -- | 2.39 | 0.243 | -- | 73.36 | 1742 | -- | -- |
| 200.3 | 0.663 | 3.16 | -0.778 | 0.0969 | -- | -0.308 | -- | 0.354 | 23 | -- | -- | -- | -- |
| 200.53 | 0.663 | 3.16 | -0.767 | 0.111 | -- | -- | -- | 0.353 | 1.02 | -- | -- | -- | -- |
| 196.38 | 0.653 | 3.17 | -0.832 | -- | -0.369 | -- | -- | 0.307 | -- | 73.96 | 1195 | -- | -- |
| 212.21 | 0.678 | 3.17 | -- | 0.152 | -0.475 | -- | -- | 0.363 | 1.35 | 88.14 | 1532 | -- | -- |
| 201.31 | 0.663 | 3.16 | -0.763 | -- | -- | -- | -- | 0.356 | -- | -- | -- | -- | -- |
| 216.06 | 0.688 | 3.14 | -- | 0.0992 | -- | -- | -- | 0.439 | 0.691 | -- | -- | -- | -- |
| 211.73 | 0.678 | 3.15 | -- | -- | -0.448 | -- | -- | 0.384 | -- | 87.31 | 1036 | -- | -- |
| 216.67 | 0.687 | 3.14 | -- | -- | -- | -- | -- | 0.44 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 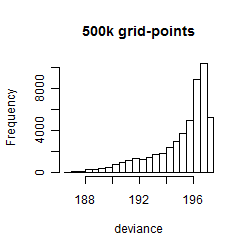
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 186.58 | humidity | 70 | 1440 | > th | dose dependent | rect. | 4 | 12 |
| 11 | 187.18 | humidity | 70 | 1440 | > th | dose dependent | sin | 1 | NA |
| 88 | 187.79 | humidity | 80 | 1440 | > th | dose independent | rect. | 1 | 19 |
| 100 | 187.84 | humidity | 80 | 1440 | > th | dose independent | rect. | 5 | 15 |
| 116 | 187.95 | humidity | 80 | 1440 | > th | dose independent | sin | 1 | NA |
| 158 | 188.14 | humidity | 80 | 1440 | > th | dose independent | rect. | 1 | 15 |
| 165 | 188.15 | humidity | 80 | 1440 | < th | dose independent | rect. | 1 | 19 |
| 173 | 188.17 | humidity | 80 | 1440 | < th | dose independent | rect. | 6 | 14 |
| 212 | 188.27 | humidity | 80 | 1440 | > th | dose independent | rect. | 5 | 11 |
| 220 | 188.27 | humidity | 60 | 1440 | > th | dose dependent | rect. | 18 | 22 |
| 281 | 188.38 | humidity | 60 | 1440 | > th | dose dependent | rect. | 11 | 23 |
| 309 | 188.43 | humidity | 80 | 1440 | < th | dose independent | sin | 1 | NA |
| 328 | 188.48 | humidity | 60 | 1440 | > th | dose dependent | no | NA | NA |
| 422 | 188.64 | humidity | 60 | 1440 | > th | dose dependent | rect. | 15 | 23 |
| 494 | 188.80 | humidity | 80 | 1440 | < th | dose independent | rect. | 1 | 15 |
| 519 | 188.86 | humidity | 80 | 1440 | < th | dose independent | rect. | 6 | 10 |
| 611 | 189.00 | humidity | 90 | 1440 | < th | dose dependent | rect. | 3 | 19 |
| 633 | 189.05 | humidity | 80 | 1440 | < th | dose independent | rect. | 8 | 8 |
| 651 | 189.08 | humidity | 60 | 1440 | > th | dose dependent | rect. | 12 | 2 |
| 669 | 189.10 | humidity | 90 | 1440 | < th | dose dependent | rect. | 12 | 22 |
| 699 | 189.17 | radiation | 10 | 1440 | > th | dose independent | rect. | 15 | 2 |
| 772 | 189.27 | humidity | 60 | 1440 | > th | dose dependent | rect. | 12 | 7 |
| 871 | 189.41 | humidity | 90 | 1440 | < th | dose dependent | rect. | 18 | 22 |
| 886 | 189.44 | humidity | 70 | 1440 | > th | dose independent | rect. | 12 | 21 |
| 915 | 189.48 | humidity | 90 | 1440 | < th | dose dependent | sin | 0 | NA |
| 964 | 189.55 | humidity | 90 | 1440 | < th | dose dependent | no | NA | NA |
| 965 | 189.55 | atmosphere | 1005 | 30 | > th | dose dependent | rect. | 18 | 1 |