Os06g0683000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Zinc finger, C2H2-type domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os06g0683000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Zinc finger, C2H2-type domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
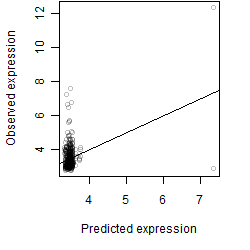 __
__
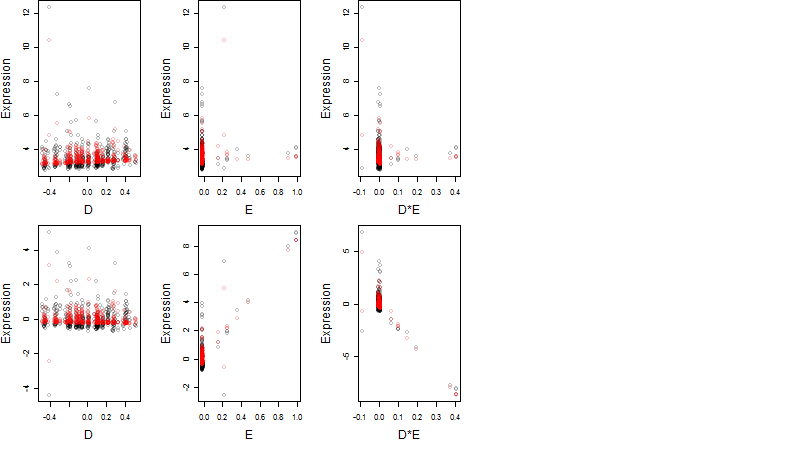
Dependence on each variable
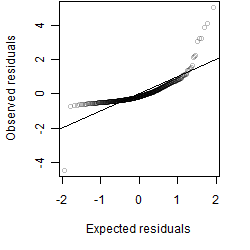
Residual plot

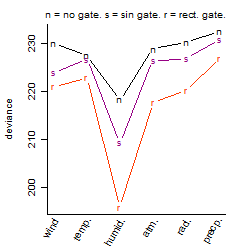
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 201.21 | 0.666 | 3.58 | -0.128 | 0.429 | 8.29 | 0.0398 | -19.3 | 0.0484 | 8.7 | 48.03 | 599 | -- | -- |
| 237.87 | 0.724 | 3.49 | 0.11 | 0.352 | -0.256 | 0.314 | -- | -0.0024 | 8.53 | 44.58 | 175 | -- | -- |
| 200.78 | 0.66 | 3.57 | -0.0995 | 0.424 | 8.35 | -- | -19.3 | 0.0485 | 8.65 | 48 | 581 | -- | -- |
| 238 | 0.719 | 3.49 | 0.119 | 0.328 | -0.19 | -- | -- | -0.0151 | 8.75 | 62 | 214 | -- | -- |
| 210.53 | 0.676 | 3.58 | -0.0798 | -- | 8.73 | -- | -20.7 | 0.0384 | -- | 47.52 | 571 | -- | -- |
| 238.06 | 0.723 | 3.49 | 0.101 | 0.349 | -- | 0.282 | -- | -0.00154 | 8.45 | -- | -- | -- | -- |
| 238.29 | 0.723 | 3.49 | 0.109 | 0.351 | -- | -- | -- | -0.00203 | 8.26 | -- | -- | -- | -- |
| 243.71 | 0.727 | 3.5 | 0.141 | -- | -0.331 | -- | -- | -0.0179 | -- | 62 | 264 | -- | -- |
| 238.37 | 0.719 | 3.49 | -- | 0.334 | -0.146 | -- | -- | -0.0239 | 8.48 | 62.61 | 214 | -- | -- |
| 245.1 | 0.732 | 3.5 | 0.111 | -- | -- | -- | -- | 0.00188 | -- | -- | -- | -- | -- |
| 238.6 | 0.723 | 3.49 | -- | 0.351 | -- | -- | -- | -0.0141 | 8.24 | -- | -- | -- | -- |
| 244.22 | 0.728 | 3.5 | -- | -- | -0.313 | -- | -- | -0.0323 | -- | 62 | 262 | -- | -- |
| 245.43 | 0.731 | 3.5 | -- | -- | -- | -- | -- | -0.0104 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance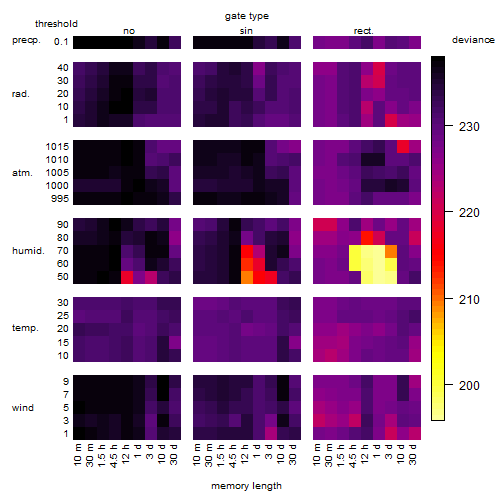
|
Histogram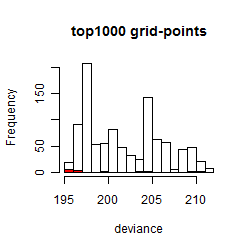 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 195.85 | humidity | 60 | 1440 | > th | dose independent | rect. | 18 | 5 |
| 3 | 195.89 | humidity | 60 | 1440 | > th | dose independent | rect. | 18 | 14 |
| 5 | 195.92 | humidity | 60 | 1440 | > th | dose independent | rect. | 18 | 1 |
| 6 | 195.93 | humidity | 60 | 1440 | > th | dose independent | rect. | 18 | 3 |
| 9 | 196.00 | humidity | 70 | 1440 | < th | dose dependent | rect. | 19 | 1 |
| 20 | 196.03 | humidity | 60 | 1440 | > th | dose independent | rect. | 18 | 9 |
| 29 | 196.37 | humidity | 60 | 1440 | < th | dose dependent | rect. | 17 | 3 |
| 73 | 196.81 | humidity | 60 | 1440 | < th | dose independent | rect. | 18 | 1 |
| 90 | 196.81 | humidity | 50 | 1440 | < th | dose independent | rect. | 17 | 17 |
| 410 | 199.34 | humidity | 50 | 4320 | < th | dose dependent | rect. | 14 | 1 |
| 590 | 203.05 | humidity | 50 | 720 | < th | dose dependent | rect. | 14 | 2 |
| 648 | 204.72 | humidity | 50 | 720 | < th | dose independent | rect. | 12 | 1 |
| 927 | 209.23 | humidity | 50 | 720 | < th | dose dependent | sin | 7 | NA |
| 992 | 210.88 | humidity | 50 | 4320 | < th | dose independent | rect. | 14 | 1 |