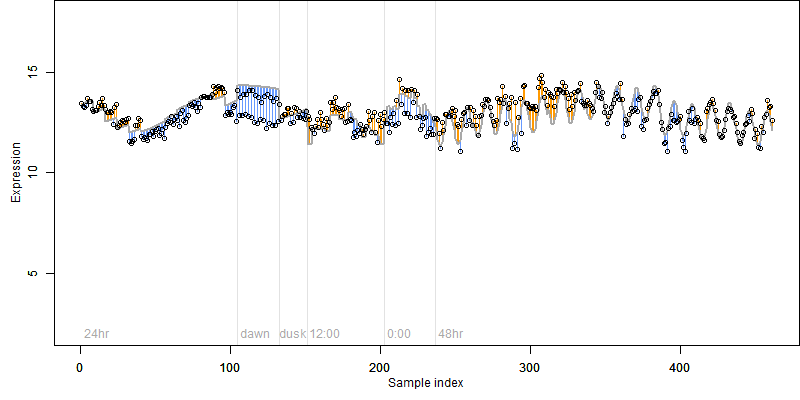
Os06g0665500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Amidase, hydantoinase/carbamoylase family protein.
FiT-DB / Search/ Help/ Sample detail
|
Os06g0665500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Amidase, hydantoinase/carbamoylase family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
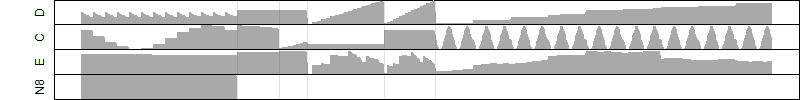
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
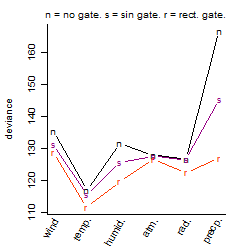
Dependence on each variable

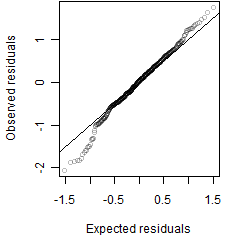
Residual plot
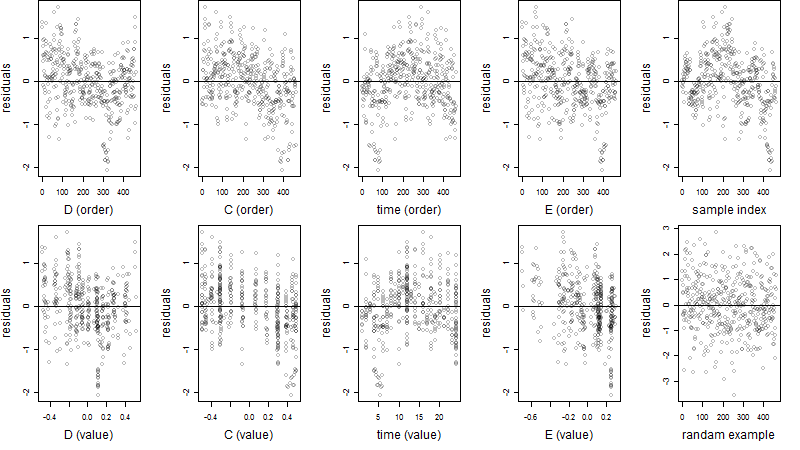
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 116.92 | 0.508 | 13.2 | -2.49 | 1.13 | 1.38 | -0.0124 | -4.81 | -0.767 | 4.2 | 20.43 | 4307 | -- | -- |
| 128.08 | 0.531 | 13 | -0.997 | 1.78 | 2.14 | 0.292 | -- | -0.444 | 3.47 | 15.17 | 4466 | -- | -- |
| 116.91 | 0.504 | 13.2 | -2.49 | 1.13 | 1.38 | -- | -4.82 | -0.764 | 4.21 | 20.5 | 4277 | -- | -- |
| 128.28 | 0.531 | 13 | -1 | 1.78 | 2.2 | -- | -- | -0.444 | 3.47 | -12.5 | 5868 | -- | -- |
| 183.77 | 0.631 | 13.2 | -2.4 | -- | 1.51 | -- | -4.46 | -0.752 | -- | 20.1 | 4080 | -- | -- |
| 174.97 | 0.62 | 13 | -0.575 | 1.19 | -- | -0.0971 | -- | -0.136 | 4.08 | -- | -- | -- | -- |
| 175 | 0.619 | 13 | -0.574 | 1.19 | -- | -- | -- | -0.135 | 4.08 | -- | -- | -- | -- |
| 194.41 | 0.649 | 13 | -1.09 | -- | 1.84 | -- | -- | -0.435 | -- | -22.56 | 1218 | -- | -- |
| 152.22 | 0.578 | 13 | -- | 1.63 | 1.69 | -- | -- | -0.276 | 3.57 | 14.8 | 1568 | -- | -- |
| 252.26 | 0.742 | 13 | -0.525 | -- | -- | -- | -- | -0.103 | -- | -- | -- | -- | -- |
| 183.68 | 0.634 | 13 | -- | 1.18 | -- | -- | -- | -0.0713 | 4.09 | -- | -- | -- | -- |
| 221.06 | 0.692 | 13 | -- | -- | 1.23 | -- | -- | -0.25 | -- | 17.3 | 1195 | -- | -- |
| 259.54 | 0.752 | 13 | -- | -- | -- | -- | -- | -0.0452 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram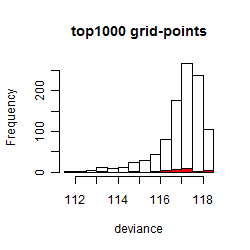 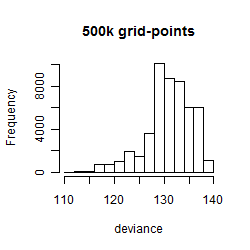
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 111.77 | temperature | 20 | 1440 | > th | dose dependent | rect. | 24 | 1 |
| 7 | 113.04 | temperature | 20 | 1440 | > th | dose dependent | rect. | 21 | 6 |
| 47 | 114.70 | temperature | 25 | 4320 | < th | dose independent | rect. | 11 | 18 |
| 53 | 114.85 | temperature | 25 | 4320 | > th | dose independent | rect. | 11 | 18 |
| 96 | 115.62 | temperature | 20 | 4320 | > th | dose dependent | rect. | 19 | 2 |
| 98 | 115.65 | temperature | 20 | 1440 | > th | dose dependent | sin | 10 | NA |
| 145 | 116.10 | temperature | 20 | 4320 | > th | dose dependent | rect. | 14 | 14 |
| 161 | 116.19 | temperature | 20 | 4320 | > th | dose dependent | sin | 15 | NA |
| 180 | 116.30 | temperature | 25 | 4320 | < th | dose independent | sin | 18 | NA |
| 223 | 116.53 | temperature | 25 | 4320 | > th | dose independent | rect. | 19 | 22 |
| 226 | 116.54 | temperature | 25 | 4320 | > th | dose independent | sin | 18 | NA |
| 250 | 116.63 | temperature | 25 | 4320 | > th | dose independent | rect. | 11 | 11 |
| 254 | 116.64 | temperature | 25 | 4320 | < th | dose independent | rect. | 19 | 22 |
| 320 | 116.84 | temperature | 20 | 4320 | > th | dose dependent | rect. | 17 | 22 |
| 326 | 116.86 | temperature | 25 | 4320 | < th | dose independent | rect. | 23 | 22 |
| 341 | 116.90 | temperature | 25 | 4320 | > th | dose independent | rect. | 23 | 22 |
| 402 | 117.03 | temperature | 20 | 4320 | > th | dose dependent | no | NA | NA |
| 411 | 117.04 | temperature | 20 | 4320 | > th | dose dependent | rect. | 23 | 23 |
| 420 | 117.06 | temperature | 25 | 4320 | > th | dose independent | rect. | 23 | 17 |
| 444 | 117.10 | temperature | 20 | 4320 | > th | dose dependent | rect. | 1 | 23 |
| 473 | 117.17 | temperature | 25 | 4320 | < th | dose independent | rect. | 23 | 17 |
| 480 | 117.18 | temperature | 25 | 4320 | < th | dose independent | no | NA | NA |
| 503 | 117.21 | temperature | 30 | 1440 | < th | dose dependent | rect. | 24 | 5 |
| 519 | 117.24 | temperature | 25 | 4320 | > th | dose independent | no | NA | NA |
| 547 | 117.30 | temperature | 30 | 1440 | < th | dose dependent | rect. | 21 | 8 |
| 726 | 117.65 | temperature | 25 | 4320 | < th | dose independent | rect. | 13 | 23 |
| 748 | 117.70 | temperature | 25 | 4320 | > th | dose independent | rect. | 13 | 23 |
| 897 | 118.01 | temperature | 30 | 1440 | < th | dose dependent | rect. | 24 | 1 |
| 917 | 118.05 | temperature | 20 | 4320 | > th | dose dependent | sin | 5 | NA |
| 942 | 118.11 | temperature | 25 | 4320 | > th | dose independent | rect. | 11 | 1 |