Os06g0347100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Acyl-coenzyme A oxidase 4, peroxisomal (EC 1.3.3.6) (AOX 4) (Short- chain acyl-CoA oxidase) (SAOX) (AtCX4) (G6p) (AtG6).


FiT-DB / Search/ Help/ Sample detail
|
Os06g0347100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Acyl-coenzyme A oxidase 4, peroxisomal (EC 1.3.3.6) (AOX 4) (Short- chain acyl-CoA oxidase) (SAOX) (AtCX4) (G6p) (AtG6).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
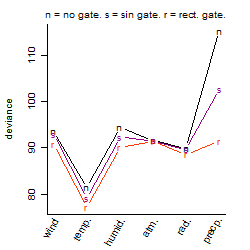
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 80.36 | 0.421 | 11.6 | -2.47 | 0.498 | 1.32 | -0.184 | -1.21 | -0.383 | 21.2 | 21.4 | 3949 | -- | -- |
| 82.66 | 0.423 | 11.6 | -2.17 | 0.503 | 1.45 | -0.258 | -- | -0.309 | 21.2 | 21.4 | 4005 | -- | -- |
| 78.74 | 0.413 | 11.6 | -2.25 | 0.445 | 1.22 | -- | -1.12 | -0.309 | 21.1 | 22.2 | 2499 | -- | -- |
| 82.53 | 0.423 | 11.6 | -1.97 | 0.452 | 1.34 | -- | -- | -0.251 | 20.9 | 21.8 | 2467 | -- | -- |
| 90.16 | 0.442 | 11.6 | -2.24 | -- | 1.26 | -- | -1.19 | -0.381 | -- | 22.24 | 2305 | -- | -- |
| 134.62 | 0.544 | 11.5 | -1.31 | 0.538 | -- | -0.186 | -- | 0.176 | 21.7 | -- | -- | -- | -- |
| 134.75 | 0.544 | 11.5 | -1.31 | 0.539 | -- | -- | -- | 0.176 | 21.7 | -- | -- | -- | -- |
| 87.46 | 0.436 | 11.6 | -1.75 | -- | 1.25 | -- | -- | -0.223 | -- | 22 | 925 | -- | -- |
| 150.26 | 0.571 | 11.5 | -- | 0.411 | 0.801 | -- | -- | 0.0712 | 20.5 | 22.32 | 979 | -- | -- |
| 152.02 | 0.576 | 11.5 | -1.3 | -- | -- | -- | -- | 0.175 | -- | -- | -- | -- | -- |
| 179.79 | 0.627 | 11.4 | -- | 0.537 | -- | -- | -- | 0.321 | 21.5 | -- | -- | -- | -- |
| 158.93 | 0.587 | 11.5 | -- | -- | 0.868 | -- | -- | 0.051 | -- | 22.4 | 882 | -- | -- |
| 196.76 | 0.655 | 11.4 | -- | -- | -- | -- | -- | 0.32 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 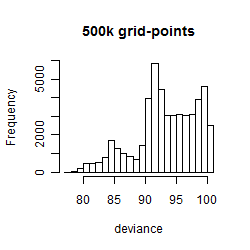
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 77.19 | temperature | 30 | 720 | < th | dose dependent | rect. | 20 | 19 |
| 11 | 78.36 | temperature | 20 | 720 | > th | dose dependent | rect. | 21 | 16 |
| 33 | 79.16 | temperature | 20 | 720 | > th | dose dependent | sin | 6 | NA |
| 35 | 79.25 | temperature | 15 | 1440 | > th | dose dependent | rect. | 18 | 16 |
| 90 | 79.56 | temperature | 15 | 1440 | > th | dose dependent | sin | 10 | NA |
| 101 | 79.62 | temperature | 30 | 1440 | < th | dose dependent | sin | 10 | NA |
| 161 | 79.82 | temperature | 30 | 270 | < th | dose dependent | rect. | 22 | 23 |
| 202 | 79.92 | temperature | 10 | 720 | > th | dose dependent | rect. | 23 | 20 |
| 210 | 79.95 | temperature | 15 | 1440 | > th | dose dependent | rect. | 1 | 9 |
| 217 | 79.96 | temperature | 30 | 270 | < th | dose dependent | rect. | 24 | 23 |
| 239 | 80.01 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 9 |
| 279 | 80.10 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 6 |
| 341 | 80.20 | temperature | 20 | 270 | > th | dose dependent | rect. | 16 | 23 |
| 369 | 80.28 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 5 |
| 375 | 80.29 | temperature | 30 | 720 | < th | dose dependent | sin | 5 | NA |
| 394 | 80.33 | temperature | 25 | 1440 | < th | dose dependent | rect. | 22 | 1 |
| 801 | 81.31 | temperature | 10 | 270 | > th | dose dependent | rect. | 5 | 23 |
| 806 | 81.34 | temperature | 30 | 270 | < th | dose dependent | no | NA | NA |
| 892 | 81.51 | temperature | 30 | 1440 | < th | dose dependent | no | NA | NA |
| 943 | 81.61 | temperature | 30 | 1440 | < th | dose dependent | rect. | 7 | 23 |
| 957 | 81.64 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 16 |