Os05g0568300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to 50S ribosomal protein L12, chloroplast precursor (CL12).


FiT-DB / Search/ Help/ Sample detail
|
Os05g0568300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to 50S ribosomal protein L12, chloroplast precursor (CL12).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
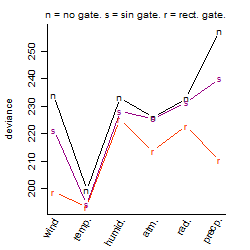
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 187.86 | 0.644 | 11.7 | 1.09 | 0.348 | -1.78 | 0.964 | -0.892 | -0.231 | 8 | 21.5 | 1392 | -- | -- |
| 188.11 | 0.639 | 11.6 | 1.2 | 0.338 | -1.59 | 1.03 | -- | -0.174 | 7.74 | 22.03 | 1358 | -- | -- |
| 190.53 | 0.643 | 11.7 | 1.1 | 0.343 | -1.81 | -- | -0.987 | -0.24 | 8.22 | 21.46 | 1374 | -- | -- |
| 190.2 | 0.642 | 11.6 | 1.15 | 0.275 | -1.51 | -- | -- | -0.164 | 6.74 | 22.3 | 1122 | -- | -- |
| 191.9 | 0.645 | 11.7 | 0.987 | -- | -1.51 | -- | -0.558 | -0.221 | -- | 22.16 | 986 | -- | -- |
| 278.64 | 0.783 | 11.8 | 0.612 | 0.358 | -- | 0.866 | -- | -0.681 | 8.51 | -- | -- | -- | -- |
| 280.9 | 0.785 | 11.8 | 0.641 | 0.355 | -- | -- | -- | -0.68 | 9.07 | -- | -- | -- | -- |
| 192.66 | 0.646 | 11.7 | 1.15 | -- | -1.48 | -- | -- | -0.176 | -- | 22.26 | 988 | -- | -- |
| 221.48 | 0.693 | 11.7 | -- | 0.233 | -1.23 | -- | -- | -0.372 | 6.38 | 22.39 | 976 | -- | -- |
| 288.14 | 0.793 | 11.8 | 0.64 | -- | -- | -- | -- | -0.678 | -- | -- | -- | -- | -- |
| 291.73 | 0.799 | 11.8 | -- | 0.355 | -- | -- | -- | -0.751 | 8.93 | -- | -- | -- | -- |
| 223.51 | 0.696 | 11.7 | -- | -- | -1.22 | -- | -- | -0.371 | -- | 22.4 | 856 | -- | -- |
| 298.96 | 0.807 | 11.8 | -- | -- | -- | -- | -- | -0.749 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 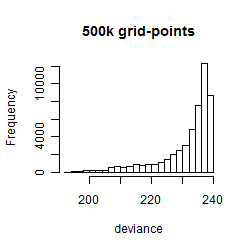
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 193.34 | temperature | 25 | 1440 | < th | dose dependent | rect. | 16 | 13 |
| 22 | 194.36 | temperature | 25 | 1440 | < th | dose dependent | sin | 14 | NA |
| 38 | 194.72 | temperature | 20 | 1440 | < th | dose independent | rect. | 5 | 1 |
| 49 | 195.08 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 1 |
| 62 | 195.53 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 4 |
| 86 | 196.09 | temperature | 20 | 1440 | > th | dose independent | rect. | 5 | 1 |
| 91 | 196.24 | temperature | 15 | 1440 | > th | dose dependent | rect. | 1 | 1 |
| 94 | 196.28 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 108 | 196.57 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 1 |
| 112 | 196.63 | temperature | 15 | 1440 | > th | dose dependent | rect. | 1 | 4 |
| 228 | 198.28 | temperature | 20 | 1440 | < th | dose independent | rect. | 4 | 3 |
| 263 | 198.87 | wind | 3 | 43200 | > th | dose independent | rect. | 1 | 1 |
| 293 | 199.21 | temperature | 25 | 720 | < th | dose dependent | rect. | 24 | 23 |
| 315 | 199.37 | temperature | 25 | 1440 | < th | dose dependent | no | NA | NA |
| 326 | 199.56 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 23 |
| 330 | 199.59 | temperature | 20 | 4320 | > th | dose independent | rect. | 22 | 1 |
| 652 | 203.05 | temperature | 20 | 4320 | < th | dose independent | rect. | 21 | 2 |
| 671 | 203.22 | temperature | 20 | 1440 | > th | dose independent | rect. | 18 | 13 |
| 695 | 203.53 | wind | 3 | 43200 | < th | dose independent | rect. | 1 | 1 |
| 762 | 204.33 | wind | 1 | 43200 | > th | dose dependent | rect. | 1 | 1 |
| 766 | 204.43 | temperature | 20 | 1440 | < th | dose independent | rect. | 18 | 13 |
| 840 | 205.04 | temperature | 20 | 4320 | < th | dose independent | rect. | 20 | 4 |
| 852 | 205.13 | wind | 9 | 14400 | > th | dose independent | rect. | 5 | 7 |
| 857 | 205.19 | wind | 9 | 14400 | < th | dose independent | rect. | 6 | 6 |
| 882 | 205.42 | temperature | 20 | 1440 | > th | dose independent | sin | 9 | NA |
| 899 | 205.60 | temperature | 20 | 4320 | < th | dose independent | rect. | 18 | 6 |
| 908 | 205.65 | temperature | 20 | 4320 | > th | dose independent | rect. | 18 | 6 |
| 938 | 205.91 | temperature | 20 | 1440 | > th | dose independent | rect. | 20 | 15 |
| 967 | 206.09 | temperature | 15 | 1440 | > th | dose dependent | sin | 11 | NA |