Os03g0850900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Chemocyanin precursor (Basic blue protein) (Plantacyanin).


FiT-DB / Search/ Help/ Sample detail
|
Os03g0850900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Chemocyanin precursor (Basic blue protein) (Plantacyanin).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
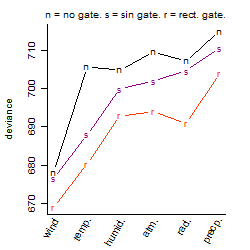
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 669.7 | 1.22 | 3.77 | -0.416 | 0.609 | 3.48 | -0.122 | 5.77 | -0.000607 | 1.76 | 8.791 | 1690 | -- | -- |
| 672.37 | 1.21 | 3.72 | -0.51 | 0.611 | 2.41 | -0.0282 | -- | 0.0319 | 1.68 | 8.498 | 1631 | -- | -- |
| 669.54 | 1.21 | 3.76 | -0.423 | 0.61 | 3.42 | -- | 5.52 | -0.0049 | 1.72 | 9 | 1690 | -- | -- |
| 667.05 | 1.2 | 3.72 | -0.641 | 0.609 | 2.44 | -- | -- | -3.29e-05 | 1.51 | 10.4 | 1665 | -- | -- |
| 691.56 | 1.22 | 3.76 | -0.368 | -- | 3.46 | -- | 6.19 | 0.008 | -- | 9 | 1838 | -- | -- |
| 728.99 | 1.27 | 3.75 | -0.974 | 0.615 | -- | -0.0692 | -- | -0.161 | 1.33 | -- | -- | -- | -- |
| 729.01 | 1.26 | 3.75 | -0.972 | 0.615 | -- | -- | -- | -0.161 | 1.31 | -- | -- | -- | -- |
| 690.4 | 1.22 | 3.71 | -0.618 | -- | 2.41 | -- | -- | 0.00486 | -- | 10.4 | 1665 | -- | -- |
| 677.47 | 1.21 | 3.7 | -- | 0.614 | 2.57 | -- | -- | 0.0717 | 1.66 | 10.37 | 1677 | -- | -- |
| 752.32 | 1.28 | 3.74 | -0.948 | -- | -- | -- | -- | -0.148 | -- | -- | -- | -- | -- |
| 753.91 | 1.28 | 3.73 | -- | 0.598 | -- | -- | -- | -0.0526 | 1.25 | -- | -- | -- | -- |
| 700.16 | 1.23 | 3.71 | -- | -- | 2.54 | -- | -- | 0.0416 | -- | 10.32 | 1665 | -- | -- |
| 776.02 | 1.3 | 3.72 | -- | -- | -- | -- | -- | -0.0429 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 669.01 | wind | 9 | 1440 | < th | dose independent | rect. | 22 | 6 |
| 2 | 671.18 | wind | 9 | 1440 | > th | dose independent | rect. | 19 | 1 |
| 3 | 671.18 | wind | 9 | 1440 | > th | dose dependent | rect. | 19 | 1 |
| 6 | 672.69 | wind | 7 | 1440 | < th | dose independent | rect. | 23 | 1 |
| 8 | 674.33 | wind | 9 | 1440 | < th | dose independent | rect. | 16 | 1 |
| 9 | 674.96 | wind | 9 | 1440 | > th | dose independent | rect. | 14 | 3 |
| 12 | 675.69 | wind | 9 | 1440 | > th | dose dependent | rect. | 15 | 2 |
| 16 | 676.09 | wind | 7 | 1440 | > th | dose dependent | rect. | 15 | 19 |
| 18 | 676.19 | wind | 9 | 1440 | < th | dose independent | rect. | 18 | 10 |
| 23 | 676.58 | wind | 9 | 1440 | < th | dose independent | sin | 11 | NA |
| 24 | 676.62 | wind | 9 | 1440 | > th | dose independent | rect. | 15 | 5 |
| 26 | 676.66 | wind | 7 | 1440 | > th | dose dependent | sin | 12 | NA |
| 29 | 676.73 | wind | 7 | 1440 | > th | dose dependent | rect. | 15 | 14 |
| 47 | 677.03 | wind | 9 | 1440 | > th | dose independent | rect. | 13 | 21 |
| 54 | 677.09 | wind | 9 | 1440 | < th | dose independent | rect. | 12 | 16 |
| 64 | 677.20 | wind | 9 | 1440 | > th | dose independent | rect. | 13 | 13 |
| 75 | 677.23 | wind | 7 | 1440 | < th | dose independent | rect. | 19 | 1 |
| 85 | 677.27 | wind | 7 | 1440 | > th | dose independent | rect. | 15 | 19 |
| 88 | 677.28 | wind | 7 | 1440 | > th | dose dependent | rect. | 21 | 23 |
| 90 | 677.30 | wind | 9 | 1440 | < th | dose independent | rect. | 12 | 22 |
| 104 | 677.39 | wind | 9 | 1440 | > th | dose dependent | rect. | 13 | 13 |
| 113 | 677.40 | wind | 9 | 1440 | > th | dose independent | sin | 11 | NA |
| 121 | 677.45 | wind | 9 | 1440 | > th | dose independent | rect. | 13 | 10 |
| 127 | 677.45 | wind | 9 | 1440 | < th | dose independent | rect. | 12 | 20 |
| 143 | 677.52 | wind | 7 | 1440 | > th | dose dependent | rect. | 15 | 9 |
| 152 | 677.57 | wind | 9 | 1440 | > th | dose independent | rect. | 18 | 5 |
| 156 | 677.57 | wind | 9 | 1440 | > th | dose dependent | rect. | 13 | 10 |
| 162 | 677.61 | wind | 9 | 1440 | < th | dose independent | rect. | 15 | 13 |
| 186 | 677.71 | wind | 9 | 1440 | > th | dose independent | sin | 15 | NA |
| 196 | 677.75 | wind | 7 | 1440 | < th | dose independent | rect. | 19 | 6 |
| 254 | 677.98 | wind | 9 | 1440 | < th | dose independent | rect. | 15 | 19 |
| 270 | 678.00 | wind | 7 | 1440 | > th | dose dependent | rect. | 1 | 23 |
| 283 | 678.07 | wind | 9 | 1440 | < th | dose independent | rect. | 15 | 17 |
| 297 | 678.17 | wind | 7 | 1440 | > th | dose dependent | no | NA | NA |
| 299 | 678.17 | wind | 7 | 1440 | > th | dose dependent | rect. | 7 | 22 |
| 322 | 678.25 | wind | 7 | 1440 | > th | dose dependent | rect. | 4 | 23 |
| 341 | 678.33 | wind | 7 | 1440 | > th | dose dependent | rect. | 18 | 23 |
| 383 | 678.51 | wind | 9 | 1440 | > th | dose independent | rect. | 21 | 23 |
| 500 | 678.95 | wind | 7 | 1440 | < th | dose independent | rect. | 17 | 16 |
| 551 | 679.13 | wind | 7 | 1440 | > th | dose dependent | rect. | 1 | 19 |
| 573 | 679.24 | wind | 7 | 1440 | > th | dose dependent | rect. | 19 | 6 |
| 619 | 679.48 | wind | 9 | 1440 | < th | dose independent | rect. | 21 | 21 |
| 645 | 679.57 | wind | 9 | 1440 | < th | dose independent | rect. | 21 | 23 |
| 653 | 679.61 | wind | 9 | 1440 | < th | dose independent | rect. | 9 | 19 |
| 657 | 679.63 | wind | 9 | 1440 | > th | dose independent | no | NA | NA |
| 658 | 679.63 | wind | 9 | 1440 | > th | dose independent | rect. | 9 | 17 |
| 673 | 679.63 | wind | 9 | 1440 | > th | dose independent | rect. | 1 | 22 |
| 722 | 679.77 | wind | 7 | 1440 | > th | dose independent | rect. | 18 | 23 |
| 725 | 679.79 | wind | 9 | 1440 | < th | dose independent | rect. | 6 | 22 |
| 753 | 679.85 | wind | 9 | 1440 | < th | dose independent | rect. | 9 | 23 |
| 758 | 679.86 | wind | 9 | 1440 | > th | dose independent | rect. | 9 | 14 |
| 798 | 679.88 | wind | 9 | 1440 | < th | dose independent | rect. | 24 | 23 |
| 880 | 680.00 | wind | 9 | 1440 | > th | dose independent | rect. | 1 | 19 |
| 949 | 680.11 | wind | 9 | 1440 | < th | dose independent | rect. | 19 | 23 |
| 958 | 680.13 | wind | 9 | 1440 | > th | dose dependent | rect. | 9 | 11 |
| 970 | 680.16 | wind | 9 | 1440 | < th | dose independent | no | NA | NA |
| 992 | 680.19 | wind | 9 | 1440 | < th | dose independent | rect. | 22 | 10 |