Os03g0816500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : 2OG-Fe(II) oxygenase domain containing protein.

FiT-DB / Search/ Help/ Sample detail
|
Os03g0816500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : 2OG-Fe(II) oxygenase domain containing protein.
|
|
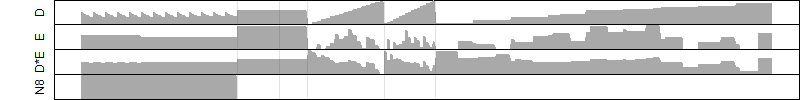
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

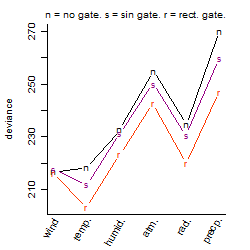
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 193.73 | 0.654 | 4.33 | -2.93 | 0.384 | 1.87 | -0.874 | -4.3 | -0.619 | 14.2 | 22.88 | 1427 | 15.4 | 0.058 |
| 221.34 | 0.693 | 4.31 | -1.76 | 0.368 | 1.82 | -0.529 | -- | -0.591 | 14 | 21.68 | 1446 | 15.8 | 0.0829 |
| 195.99 | 0.652 | 4.33 | -2.9 | 0.385 | 1.87 | -- | -4.03 | -0.61 | 14.5 | 22.84 | 1440 | 15.5 | 0.0165 |
| 214.65 | 0.682 | 4.2 | -1.89 | 0.281 | 1.87 | -- | -- | -0.427 | 13.8 | 20.36 | 1441 | 15.9 | 0.138 |
| 204.7 | 0.666 | 4.33 | -2.91 | -- | 1.82 | -- | -4.11 | -0.604 | -- | 22.88 | 1440 | 15.5 | 0.0167 |
| 303.09 | 0.816 | 4.15 | -1.25 | 0.307 | -- | -0.767 | -- | -0.173 | 14.3 | -- | -- | -- | -- |
| 305.23 | 0.818 | 4.15 | -1.27 | 0.309 | -- | -- | -- | -0.178 | 14.6 | -- | -- | -- | -- |
| 221.08 | 0.693 | 4.17 | -1.73 | -- | 1.91 | -- | -- | -0.316 | -- | 22.38 | 1650 | 15.9 | 0.143 |
| 284.59 | 0.786 | 4.12 | -- | 0.318 | 1.85 | -- | -- | -0.114 | 13.4 | 22.6 | 1577 | 16.6 | 0.0166 |
| 310.82 | 0.824 | 4.15 | -1.28 | -- | -- | -- | -- | -0.185 | -- | -- | -- | -- | -- |
| 347.82 | 0.872 | 4.12 | -- | 0.334 | -- | -- | -- | -0.036 | 14.6 | -- | -- | -- | -- |
| 290.21 | 0.793 | 4.13 | -- | -- | 1.76 | -- | -- | -0.124 | -- | 22.2 | 1585 | 16.6 | 0.000141 |
| 354.34 | 0.879 | 4.12 | -- | -- | -- | -- | -- | -0.0429 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 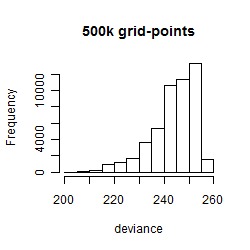
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 203.29 | temperature | 25 | 1440 | > th | dose dependent | rect. | 15 | 1 |
| 4 | 204.34 | temperature | 20 | 1440 | > th | dose dependent | rect. | 13 | 5 |
| 102 | 211.94 | temperature | 20 | 1440 | > th | dose dependent | sin | 20 | NA |
| 144 | 213.08 | temperature | 25 | 1440 | > th | dose dependent | rect. | 13 | 19 |
| 319 | 216.17 | wind | 7 | 43200 | < th | dose independent | rect. | 11 | 23 |
| 325 | 216.24 | wind | 7 | 43200 | > th | dose independent | rect. | 11 | 23 |
| 327 | 216.26 | wind | 7 | 43200 | > th | dose independent | rect. | 13 | 23 |
| 328 | 216.26 | wind | 7 | 43200 | < th | dose independent | rect. | 13 | 23 |
| 381 | 216.66 | wind | 7 | 43200 | < th | dose independent | rect. | 9 | 23 |
| 384 | 216.68 | wind | 7 | 43200 | < th | dose independent | rect. | 18 | 23 |
| 385 | 216.69 | wind | 7 | 43200 | < th | dose independent | rect. | 3 | 23 |
| 386 | 216.70 | wind | 7 | 43200 | < th | dose independent | no | NA | NA |
| 390 | 216.71 | wind | 7 | 43200 | > th | dose independent | rect. | 18 | 23 |
| 394 | 216.73 | wind | 7 | 43200 | > th | dose independent | no | NA | NA |
| 395 | 216.73 | wind | 7 | 43200 | > th | dose independent | rect. | 9 | 23 |
| 401 | 216.75 | wind | 7 | 43200 | > th | dose independent | rect. | 2 | 23 |
| 423 | 216.79 | wind | 7 | 43200 | < th | dose independent | rect. | 23 | 23 |
| 435 | 216.84 | wind | 7 | 43200 | > th | dose independent | rect. | 23 | 23 |
| 583 | 217.36 | wind | 7 | 43200 | > th | dose dependent | rect. | 13 | 19 |
| 643 | 217.62 | wind | 7 | 43200 | > th | dose independent | sin | 15 | NA |
| 657 | 217.70 | wind | 7 | 43200 | < th | dose independent | sin | 16 | NA |
| 663 | 217.71 | wind | 5 | 43200 | > th | dose dependent | rect. | 11 | 19 |
| 781 | 218.15 | wind | 7 | 43200 | > th | dose dependent | sin | 15 | NA |
| 824 | 218.35 | temperature | 25 | 1440 | > th | dose dependent | no | NA | NA |
| 950 | 218.91 | wind | 5 | 43200 | > th | dose dependent | rect. | 4 | 21 |