Os03g0652100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : CCR4-NOT complex, subunits 3 and 5 family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os03g0652100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : CCR4-NOT complex, subunits 3 and 5 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
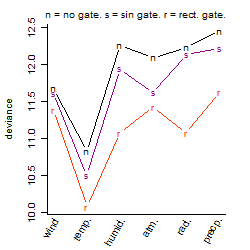
Dependence on each variable

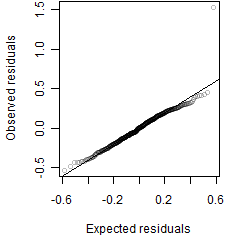
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10.82 | 0.155 | 11.5 | -0.128 | 0.327 | 0.526 | 0.137 | -0.179 | -0.00931 | 12 | 15.4 | 714 | -- | -- |
| 10.84 | 0.153 | 11.5 | -0.0964 | 0.325 | 0.527 | 0.16 | -- | 0.00071 | 12 | 14.9 | 719 | -- | -- |
| 10.89 | 0.154 | 11.5 | -0.14 | 0.324 | 0.524 | -- | -0.275 | -0.0163 | 11.9 | 15.77 | 684 | -- | -- |
| 10.95 | 0.154 | 11.5 | -0.0885 | 0.326 | 0.527 | -- | -- | 0.000822 | 12.1 | 15.01 | 716 | -- | -- |
| 11.54 | 0.159 | 11.5 | -0.105 | -- | 0.676 | -- | -0.0739 | -0.0217 | -- | 15.77 | 684 | -- | -- |
| 14.49 | 0.178 | 11.4 | 0.0414 | 0.27 | -- | 0.128 | -- | 0.0825 | 13.5 | -- | -- | -- | -- |
| 14.56 | 0.179 | 11.4 | 0.0457 | 0.271 | -- | -- | -- | 0.0832 | 13.5 | -- | -- | -- | -- |
| 11.55 | 0.159 | 11.5 | -0.0939 | -- | 0.674 | -- | -- | -0.0187 | -- | 15.77 | 684 | -- | -- |
| 11.13 | 0.155 | 11.5 | -- | 0.319 | 0.482 | -- | -- | 0.0149 | 12.1 | 15.5 | 694 | -- | -- |
| 19.04 | 0.204 | 11.4 | 0.0349 | -- | -- | -- | -- | 0.0774 | -- | -- | -- | -- | -- |
| 14.61 | 0.179 | 11.4 | -- | 0.27 | -- | -- | -- | 0.0781 | 13.5 | -- | -- | -- | -- |
| 11.77 | 0.16 | 11.5 | -- | -- | 0.647 | -- | -- | -0.005 | -- | 15.77 | 684 | -- | -- |
| 19.07 | 0.204 | 11.4 | -- | -- | -- | -- | -- | 0.0735 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance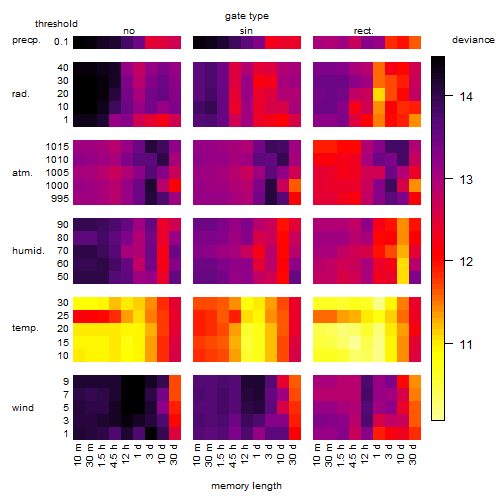
|
Histogram 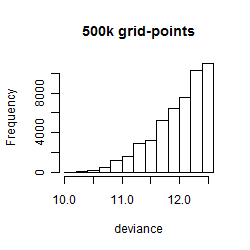
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10.07 | temperature | 15 | 270 | > th | dose dependent | rect. | 4 | 17 |
| 2 | 10.14 | temperature | 10 | 1440 | > th | dose dependent | rect. | 8 | 1 |
| 5 | 10.18 | temperature | 30 | 1440 | < th | dose dependent | rect. | 8 | 1 |
| 20 | 10.27 | temperature | 15 | 270 | > th | dose dependent | rect. | 1 | 21 |
| 37 | 10.32 | temperature | 10 | 270 | > th | dose dependent | rect. | 12 | 23 |
| 44 | 10.33 | temperature | 15 | 90 | > th | dose dependent | rect. | 7 | 15 |
| 122 | 10.47 | temperature | 15 | 30 | > th | dose dependent | rect. | 3 | 23 |
| 139 | 10.50 | temperature | 15 | 720 | > th | dose dependent | sin | 2 | NA |
| 150 | 10.51 | temperature | 10 | 720 | > th | dose dependent | rect. | 22 | 21 |
| 219 | 10.59 | temperature | 30 | 90 | < th | dose dependent | rect. | 9 | 22 |
| 286 | 10.64 | temperature | 30 | 10 | < th | dose dependent | rect. | 21 | 23 |
| 300 | 10.65 | temperature | 30 | 720 | < th | dose dependent | rect. | 2 | 19 |
| 303 | 10.65 | temperature | 30 | 10 | < th | dose dependent | rect. | 9 | 23 |
| 411 | 10.72 | temperature | 20 | 30 | > th | dose dependent | rect. | 7 | 23 |
| 412 | 10.72 | temperature | 30 | 10 | < th | dose dependent | rect. | 19 | 23 |
| 447 | 10.74 | temperature | 20 | 1440 | > th | dose dependent | rect. | 7 | 13 |
| 502 | 10.75 | temperature | 30 | 10 | < th | dose dependent | rect. | 17 | 23 |
| 630 | 10.79 | temperature | 30 | 720 | < th | dose dependent | sin | 2 | NA |
| 724 | 10.80 | temperature | 15 | 10 | > th | dose dependent | rect. | 1 | 23 |
| 727 | 10.80 | temperature | 15 | 720 | > th | dose dependent | rect. | 16 | 23 |
| 730 | 10.80 | temperature | 30 | 10 | < th | dose dependent | rect. | 11 | 23 |
| 732 | 10.81 | temperature | 20 | 1440 | > th | dose dependent | rect. | 18 | 1 |
| 780 | 10.82 | temperature | 15 | 720 | > th | dose dependent | no | NA | NA |
| 819 | 10.82 | temperature | 15 | 720 | > th | dose dependent | rect. | 18 | 23 |
| 881 | 10.84 | temperature | 30 | 10 | < th | dose dependent | no | NA | NA |
| 913 | 10.84 | temperature | 30 | 720 | < th | dose dependent | rect. | 17 | 21 |
| 969 | 10.85 | temperature | 15 | 720 | > th | dose dependent | rect. | 7 | 23 |