Os03g0305600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Mitochondrial import inner membrane translocase, subunit Tim17/22 family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os03g0305600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Mitochondrial import inner membrane translocase, subunit Tim17/22 family protein.
|
|
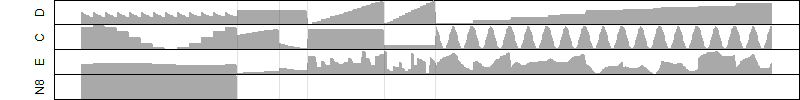
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
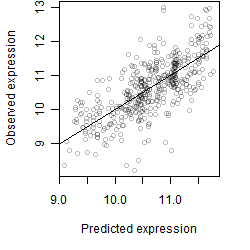
Dependence on each variable
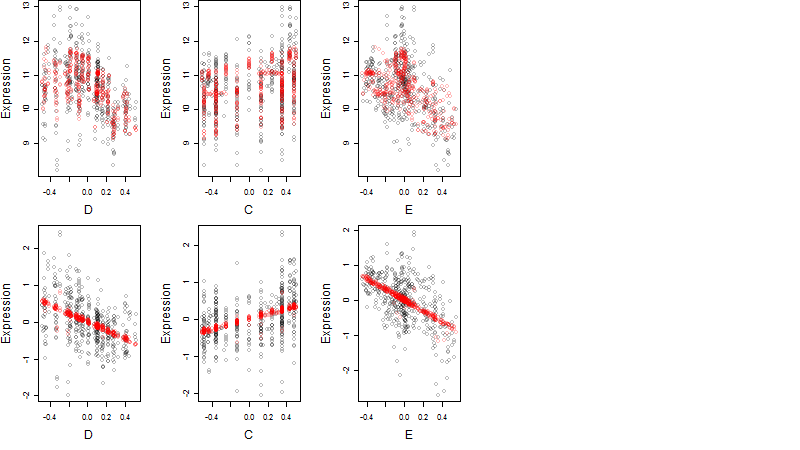
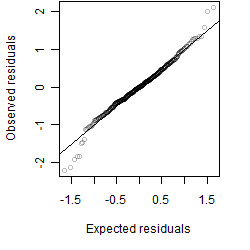
Residual plot

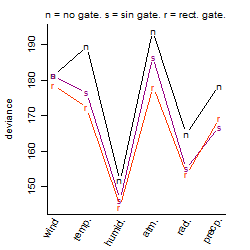
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 151.49 | 0.578 | 10.5 | -1.32 | 0.643 | -1.49 | 0.719 | 1.6 | 0.67 | 6.98 | 47.46 | 1488 | -- | -- |
| 154.43 | 0.579 | 10.5 | -1.15 | 0.667 | -1.52 | 0.0455 | -- | 0.689 | 8.92 | 44.94 | 1307 | -- | -- |
| 152.06 | 0.574 | 10.5 | -1.28 | 0.623 | -1.51 | -- | 1.43 | 0.683 | 7.89 | 42.79 | 1405 | -- | -- |
| 154.3 | 0.579 | 10.5 | -1.13 | 0.686 | -1.53 | -- | -- | 0.702 | 9.01 | 49.61 | 1281 | -- | -- |
| 172.49 | 0.612 | 10.5 | -1.3 | -- | -1.56 | -- | 1.47 | 0.678 | -- | 43.65 | 1405 | -- | -- |
| 205.37 | 0.672 | 10.5 | -1.06 | 0.648 | -- | 0.687 | -- | 0.762 | 7.08 | -- | -- | -- | -- |
| 206.2 | 0.672 | 10.5 | -1.04 | 0.65 | -- | -- | -- | 0.762 | 7.56 | -- | -- | -- | -- |
| 175.92 | 0.618 | 10.5 | -1.11 | -- | -1.46 | -- | -- | 0.702 | -- | 53.04 | 1398 | -- | -- |
| 185.52 | 0.634 | 10.5 | -- | 0.696 | -1.72 | -- | -- | 0.705 | 9.16 | 65.87 | 1268 | -- | -- |
| 228.83 | 0.707 | 10.5 | -1.04 | -- | -- | -- | -- | 0.772 | -- | -- | -- | -- | -- |
| 234.96 | 0.717 | 10.4 | -- | 0.641 | -- | -- | -- | 0.879 | 7.67 | -- | -- | -- | -- |
| 205.49 | 0.668 | 10.5 | -- | -- | -1.91 | -- | -- | 0.672 | -- | 72.44 | 1412 | -- | -- |
| 257.09 | 0.748 | 10.4 | -- | -- | -- | -- | -- | 0.887 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 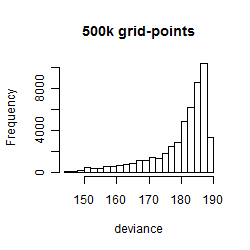
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 144.16 | humidity | 90 | 720 | > th | dose dependent | rect. | 20 | 3 |
| 2 | 144.37 | humidity | 80 | 1440 | > th | dose independent | rect. | 7 | 12 |
| 9 | 145.11 | humidity | 90 | 720 | > th | dose dependent | rect. | 8 | 16 |
| 10 | 145.14 | humidity | 80 | 1440 | < th | dose independent | rect. | 7 | 12 |
| 11 | 145.19 | humidity | 90 | 720 | > th | dose independent | rect. | 20 | 3 |
| 35 | 146.20 | humidity | 80 | 1440 | > th | dose independent | sin | 0 | NA |
| 57 | 146.93 | humidity | 80 | 1440 | < th | dose independent | sin | 0 | NA |
| 79 | 147.47 | humidity | 90 | 720 | > th | dose dependent | rect. | 15 | 9 |
| 83 | 147.55 | humidity | 70 | 1440 | > th | dose dependent | sin | 23 | NA |
| 87 | 147.62 | humidity | 70 | 1440 | > th | dose dependent | rect. | 8 | 11 |
| 105 | 147.96 | humidity | 70 | 1440 | > th | dose dependent | rect. | 5 | 14 |
| 228 | 149.75 | humidity | 90 | 720 | > th | dose dependent | sin | 20 | NA |
| 242 | 149.82 | humidity | 70 | 1440 | > th | dose dependent | rect. | 10 | 4 |
| 245 | 149.82 | humidity | 50 | 1440 | > th | dose dependent | rect. | 4 | 19 |
| 288 | 150.14 | humidity | 90 | 1440 | < th | dose dependent | rect. | 9 | 10 |
| 298 | 150.17 | humidity | 90 | 1440 | < th | dose dependent | rect. | 11 | 2 |
| 303 | 150.19 | humidity | 60 | 1440 | > th | dose dependent | rect. | 11 | 2 |
| 311 | 150.22 | humidity | 90 | 1440 | < th | dose dependent | rect. | 4 | 15 |
| 340 | 150.36 | humidity | 90 | 1440 | < th | dose dependent | sin | 0 | NA |
| 512 | 151.15 | humidity | 50 | 1440 | > th | dose dependent | rect. | 9 | 22 |
| 586 | 151.45 | humidity | 50 | 1440 | > th | dose dependent | rect. | 10 | 5 |
| 614 | 151.56 | humidity | 50 | 1440 | > th | dose dependent | no | NA | NA |
| 617 | 151.59 | humidity | 90 | 1440 | < th | dose dependent | rect. | 10 | 5 |
| 623 | 151.61 | humidity | 60 | 720 | > th | dose dependent | rect. | 17 | 6 |
| 700 | 151.93 | humidity | 90 | 720 | > th | dose independent | rect. | 7 | 17 |
| 704 | 151.93 | humidity | 50 | 1440 | > th | dose dependent | rect. | 15 | 23 |
| 731 | 152.09 | humidity | 90 | 720 | > th | dose independent | rect. | 9 | 16 |
| 812 | 152.36 | humidity | 90 | 1440 | < th | dose dependent | rect. | 9 | 22 |
| 884 | 152.68 | humidity | 90 | 1440 | < th | dose dependent | no | NA | NA |
| 941 | 152.96 | humidity | 90 | 1440 | < th | dose dependent | rect. | 15 | 23 |