Os03g0228400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Adapter-related protein complex 3 sigma 2 subunit (Sigma-adaptin 3b) (AP-3 complex sigma-3B subunit) (Sigma-3B-adaptin).

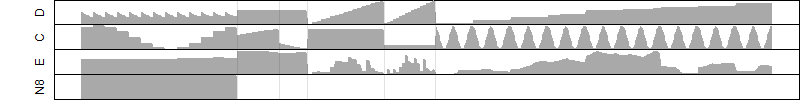
FiT-DB / Search/ Help/ Sample detail
|
Os03g0228400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Adapter-related protein complex 3 sigma 2 subunit (Sigma-adaptin 3b) (AP-3 complex sigma-3B subunit) (Sigma-3B-adaptin).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
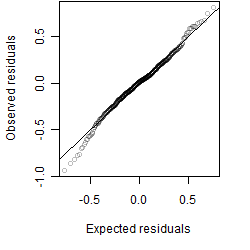
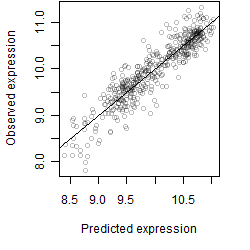 __
__

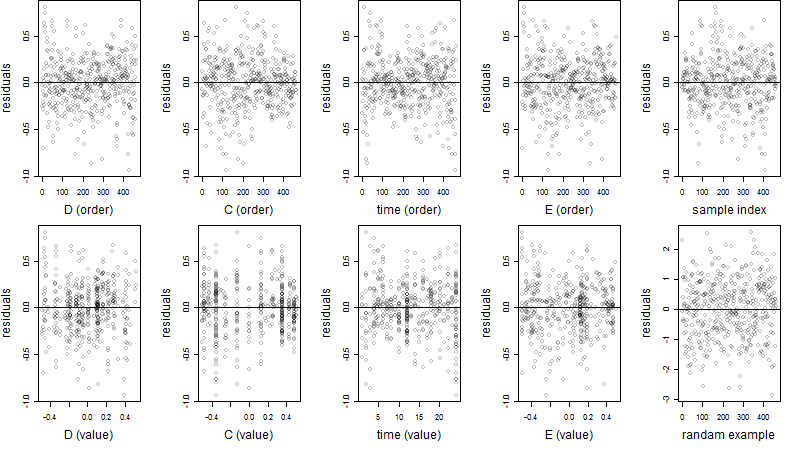
Dependence on each variable

Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 32.09 | 0.266 | 9.97 | -0.987 | 1.55 | 0.985 | 0.437 | 0.549 | -0.211 | 9.09 | 20.57 | 1337 | -- | -- |
| 32.3 | 0.265 | 9.98 | -1.1 | 1.66 | 1.03 | 0.559 | -- | -0.21 | 9.22 | 21.48 | 1128 | -- | -- |
| 32.72 | 0.266 | 9.97 | -1 | 1.54 | 0.967 | -- | 0.446 | -0.216 | 9.03 | 20.78 | 1372 | -- | -- |
| 32.87 | 0.267 | 9.99 | -1.12 | 1.55 | 0.987 | -- | -- | -0.245 | 9.04 | 21.28 | 1343 | -- | -- |
| 166.02 | 0.603 | 10 | -0.858 | -- | 1.11 | -- | 0.785 | -0.155 | -- | 20.35 | 1478 | -- | -- |
| 65.2 | 0.379 | 9.94 | -0.782 | 1.53 | -- | 0.351 | -- | -0.0206 | 8.79 | -- | -- | -- | -- |
| 65.61 | 0.379 | 9.94 | -0.771 | 1.53 | -- | -- | -- | -0.0207 | 8.77 | -- | -- | -- | -- |
| 166.64 | 0.604 | 10 | -0.991 | -- | 1.16 | -- | -- | -0.194 | -- | 20.35 | 1478 | -- | -- |
| 62.34 | 0.368 | 9.94 | -- | 1.58 | 0.717 | -- | -- | -0.0541 | 9.11 | 22.3 | 1216 | -- | -- |
| 198.05 | 0.658 | 9.98 | -0.769 | -- | -- | -- | -- | -0.00871 | -- | -- | -- | -- | -- |
| 81.26 | 0.422 | 9.92 | -- | 1.53 | -- | -- | -- | 0.0652 | 8.81 | -- | -- | -- | -- |
| 191.53 | 0.647 | 9.99 | -- | -- | 0.949 | -- | -- | -0.0556 | -- | 20.35 | 1478 | -- | -- |
| 213.63 | 0.682 | 9.96 | -- | -- | -- | -- | -- | 0.0766 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 29.54 | temperature | 30 | 90 | < th | dose dependent | rect. | 5 | 23 |
| 2 | 30.39 | temperature | 10 | 270 | > th | dose dependent | rect. | 24 | 23 |
| 3 | 30.82 | temperature | 30 | 10 | < th | dose dependent | rect. | 6 | 22 |
| 7 | 31.44 | temperature | 20 | 720 | > th | dose dependent | rect. | 17 | 21 |
| 15 | 31.97 | temperature | 10 | 720 | > th | dose dependent | rect. | 17 | 22 |
| 17 | 31.99 | temperature | 10 | 720 | > th | dose dependent | rect. | 7 | 23 |
| 22 | 32.06 | temperature | 20 | 1440 | > th | dose dependent | sin | 15 | NA |
| 24 | 32.09 | temperature | 30 | 90 | < th | dose dependent | rect. | 7 | 20 |
| 25 | 32.09 | temperature | 20 | 1440 | > th | dose dependent | rect. | 10 | 21 |
| 26 | 32.11 | temperature | 20 | 1440 | > th | dose dependent | rect. | 13 | 19 |
| 39 | 32.15 | temperature | 20 | 1440 | > th | dose dependent | rect. | 13 | 16 |
| 70 | 32.21 | temperature | 20 | 1440 | > th | dose dependent | rect. | 15 | 11 |
| 115 | 32.27 | temperature | 10 | 720 | > th | dose dependent | rect. | 15 | 23 |
| 116 | 32.28 | temperature | 20 | 1440 | > th | dose dependent | no | NA | NA |
| 153 | 32.34 | temperature | 20 | 1440 | > th | dose dependent | rect. | 1 | 23 |
| 154 | 32.34 | temperature | 20 | 1440 | > th | dose dependent | rect. | 3 | 23 |
| 156 | 32.34 | temperature | 15 | 270 | > th | dose dependent | rect. | 10 | 23 |
| 206 | 32.45 | temperature | 20 | 270 | > th | dose dependent | rect. | 12 | 23 |
| 223 | 32.50 | temperature | 10 | 1440 | > th | dose dependent | rect. | 15 | 9 |
| 240 | 32.54 | temperature | 10 | 1440 | > th | dose dependent | sin | 17 | NA |
| 249 | 32.55 | temperature | 10 | 1440 | > th | dose dependent | rect. | 10 | 16 |
| 260 | 32.57 | temperature | 10 | 270 | > th | dose dependent | no | NA | NA |
| 289 | 32.60 | temperature | 10 | 1440 | > th | dose dependent | rect. | 13 | 13 |
| 411 | 32.74 | temperature | 20 | 1440 | > th | dose dependent | sin | 7 | NA |
| 492 | 32.79 | temperature | 10 | 1440 | > th | dose dependent | rect. | 13 | 23 |
| 564 | 32.86 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 23 |
| 704 | 33.01 | temperature | 30 | 1440 | < th | dose dependent | rect. | 18 | 6 |