Os02g0694700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : RWD domain containing protein.

FiT-DB / Search/ Help/ Sample detail
|
Os02g0694700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : RWD domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

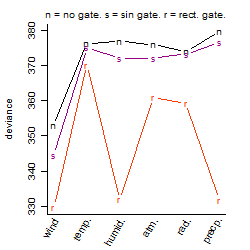
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 327.98 | 0.851 | 2.99 | 0.352 | 0.243 | 5.01 | 0.377 | 20.2 | -0.177 | 2.43 | 2.976 | 665 | 4.05 | 0.931 |
| 362.78 | 0.887 | 2.92 | 0.257 | 0.317 | 2.91 | 0.542 | -- | -0.0722 | 1.23 | 2.905 | 489 | 4.92 | 1.55 |
| 327.19 | 0.842 | 2.98 | 0.357 | 0.246 | 4.96 | -- | 19.8 | -0.178 | 2.59 | 2.9 | 652 | 4.14 | 0.768 |
| 358.26 | 0.882 | 2.91 | 0.324 | 0.248 | 2.52 | -- | -- | -0.103 | 1.27 | 3.402 | 463 | 5.02 | 1.63 |
| 329.01 | 0.845 | 2.97 | 0.313 | -- | 5.11 | -- | 20.7 | -0.15 | -- | 2.926 | 562 | 4.25 | 0.533 |
| 382.25 | 0.917 | 2.96 | 0.15 | 0.183 | -- | -0.0215 | -- | -0.205 | 5.54 | -- | -- | -- | -- |
| 382.25 | 0.916 | 2.96 | 0.15 | 0.183 | -- | -- | -- | -0.205 | 5.55 | -- | -- | -- | -- |
| 356.8 | 0.88 | 2.94 | 0.189 | -- | 2.48 | -- | -- | -0.12 | -- | 3.791 | 429 | 5.45 | 1.38 |
| 355.14 | 0.878 | 2.94 | -- | 0.171 | 2.29 | -- | -- | -0.158 | 1.29 | 3.667 | 438 | 5.43 | 1.73 |
| 383.98 | 0.916 | 2.96 | 0.156 | -- | -- | -- | -- | -0.201 | -- | -- | -- | -- | -- |
| 382.84 | 0.915 | 2.96 | -- | 0.185 | -- | -- | -- | -0.222 | 5.52 | -- | -- | -- | -- |
| 356.9 | 0.88 | 2.95 | -- | -- | 2.32 | -- | -- | -0.163 | -- | 3.9 | 446 | 5.42 | 1.77 |
| 384.62 | 0.915 | 2.96 | -- | -- | -- | -- | -- | -0.218 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 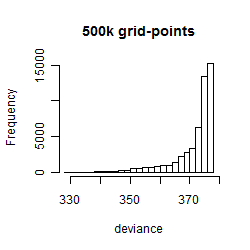
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 329.70 | wind | 3.0 | 720 | > th | dose dependent | rect. | 4 | 1 |
| 3 | 330.52 | wind | 5.0 | 720 | > th | dose independent | rect. | 4 | 1 |
| 11 | 331.22 | wind | 5.0 | 720 | > th | dose independent | rect. | 4 | 4 |
| 15 | 331.71 | precipitation | 0.1 | 1440 | > th | dose independent | rect. | 15 | 1 |
| 16 | 331.71 | precipitation | 0.1 | 1440 | > th | dose dependent | rect. | 15 | 1 |
| 17 | 331.78 | wind | 5.0 | 720 | > th | dose dependent | rect. | 2 | 6 |
| 19 | 331.87 | humidity | 90.0 | 1440 | > th | dose dependent | rect. | 15 | 1 |
| 25 | 332.55 | precipitation | 0.1 | 1440 | < th | dose independent | rect. | 15 | 1 |
| 26 | 332.55 | precipitation | 0.1 | 1440 | < th | dose dependent | rect. | 15 | 1 |
| 32 | 335.50 | wind | 9.0 | 90 | > th | dose dependent | rect. | 10 | 1 |
| 35 | 335.80 | wind | 9.0 | 90 | > th | dose independent | rect. | 10 | 1 |
| 70 | 337.91 | humidity | 90.0 | 1440 | > th | dose independent | rect. | 15 | 2 |
| 72 | 338.10 | humidity | 90.0 | 1440 | > th | dose dependent | rect. | 10 | 7 |
| 84 | 339.01 | wind | 7.0 | 10 | > th | dose dependent | rect. | 11 | 1 |
| 101 | 340.06 | wind | 9.0 | 10 | > th | dose independent | rect. | 11 | 1 |
| 272 | 344.53 | wind | 9.0 | 90 | < th | dose independent | sin | 7 | NA |
| 297 | 344.79 | wind | 9.0 | 90 | > th | dose independent | sin | 7 | NA |
| 367 | 346.03 | wind | 5.0 | 1440 | < th | dose independent | rect. | 4 | 1 |
| 382 | 346.21 | wind | 7.0 | 90 | > th | dose dependent | sin | 6 | NA |
| 768 | 349.52 | wind | 7.0 | 10 | > th | dose dependent | sin | 5 | NA |
| 984 | 350.81 | wind | 9.0 | 1440 | < th | dose independent | rect. | 10 | 1 |