Os02g0612300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Nuclear cap binding protein subunit 2 (20 kDa nuclear cap binding protein) (NCBP 20 kDa subunit) (CBP20).


FiT-DB / Search/ Help/ Sample detail
|
Os02g0612300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Nuclear cap binding protein subunit 2 (20 kDa nuclear cap binding protein) (NCBP 20 kDa subunit) (CBP20).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
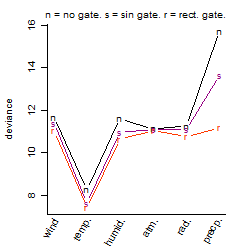
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8.21 | 0.135 | 12.2 | -0.574 | 0.134 | 0.611 | 0.108 | -0.109 | -0.142 | 21.3 | 21.41 | 2626 | -- | -- |
| 7.69 | 0.129 | 12.2 | -0.458 | 0.141 | 0.557 | -0.0013 | -- | -0.129 | 22 | 22.1 | 1406 | -- | -- |
| 8.19 | 0.133 | 12.2 | -0.571 | 0.128 | 0.594 | -- | -0.134 | -0.151 | 21.6 | 21.8 | 2606 | -- | -- |
| 7.66 | 0.129 | 12.2 | -0.451 | 0.151 | 0.569 | -- | -- | -0.129 | 21.8 | 21.9 | 1406 | -- | -- |
| 8.99 | 0.14 | 12.2 | -0.544 | -- | 0.596 | -- | -0.0825 | -0.142 | -- | 21.8 | 2263 | -- | -- |
| 18.67 | 0.203 | 12.1 | -0.233 | 0.162 | -- | 0.0359 | -- | 0.0573 | 22.4 | -- | -- | -- | -- |
| 18.67 | 0.202 | 12.1 | -0.235 | 0.161 | -- | -- | -- | 0.0571 | 22.5 | -- | -- | -- | -- |
| 8.8 | 0.138 | 12.2 | -0.422 | -- | 0.538 | -- | -- | -0.125 | -- | 22.21 | 1226 | -- | -- |
| 12.02 | 0.161 | 12.2 | -- | 0.148 | 0.437 | -- | -- | -0.0502 | 21.6 | 22.35 | 1383 | -- | -- |
| 20.27 | 0.21 | 12.1 | -0.232 | -- | -- | -- | -- | 0.0578 | -- | -- | -- | -- | -- |
| 20.12 | 0.21 | 12.1 | -- | 0.16 | -- | -- | -- | 0.0833 | 22.4 | -- | -- | -- | -- |
| 13 | 0.168 | 12.2 | -- | -- | 0.433 | -- | -- | -0.0482 | -- | 22.4 | 1096 | -- | -- |
| 21.69 | 0.217 | 12.1 | -- | -- | -- | -- | -- | 0.0835 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 7.44 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 8 |
| 30 | 7.58 | temperature | 15 | 1440 | > th | dose dependent | rect. | 21 | 8 |
| 35 | 7.60 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 2 |
| 54 | 7.63 | temperature | 25 | 1440 | < th | dose dependent | sin | 11 | NA |
| 218 | 7.83 | temperature | 15 | 1440 | > th | dose dependent | rect. | 1 | 2 |
| 341 | 7.96 | temperature | 15 | 1440 | > th | dose dependent | sin | 10 | NA |
| 495 | 8.13 | temperature | 30 | 270 | < th | dose dependent | rect. | 8 | 23 |
| 592 | 8.26 | temperature | 30 | 270 | < th | dose dependent | no | NA | NA |
| 671 | 8.32 | temperature | 30 | 270 | < th | dose dependent | rect. | 12 | 23 |
| 681 | 8.33 | temperature | 30 | 270 | < th | dose dependent | rect. | 14 | 23 |
| 683 | 8.34 | temperature | 25 | 1440 | < th | dose dependent | no | NA | NA |
| 712 | 8.36 | temperature | 30 | 270 | < th | dose dependent | rect. | 19 | 23 |
| 719 | 8.37 | temperature | 30 | 270 | < th | dose dependent | rect. | 10 | 23 |
| 765 | 8.41 | temperature | 25 | 1440 | < th | dose dependent | rect. | 4 | 23 |
| 767 | 8.41 | temperature | 25 | 1440 | < th | dose dependent | rect. | 23 | 23 |
| 821 | 8.46 | temperature | 30 | 10 | < th | dose dependent | rect. | 9 | 23 |
| 831 | 8.48 | temperature | 15 | 270 | > th | dose dependent | no | NA | NA |
| 843 | 8.48 | temperature | 30 | 10 | < th | dose dependent | rect. | 19 | 23 |
| 915 | 8.54 | temperature | 30 | 10 | < th | dose dependent | rect. | 11 | 23 |
| 932 | 8.56 | temperature | 30 | 10 | < th | dose dependent | no | NA | NA |
| 945 | 8.56 | temperature | 30 | 10 | < th | dose dependent | rect. | 15 | 23 |
| 969 | 8.58 | temperature | 15 | 270 | > th | dose dependent | rect. | 12 | 23 |