Os02g0477700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Type II inositol-1,4,5-trisphosphate 5-phosphatase 12 (EC 3.1.3.36) (At5PTase12) (FRAGILE FIBER3 protein).

FiT-DB / Search/ Help/ Sample detail
|
Os02g0477700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Type II inositol-1,4,5-trisphosphate 5-phosphatase 12 (EC 3.1.3.36) (At5PTase12) (FRAGILE FIBER3 protein).
|
|
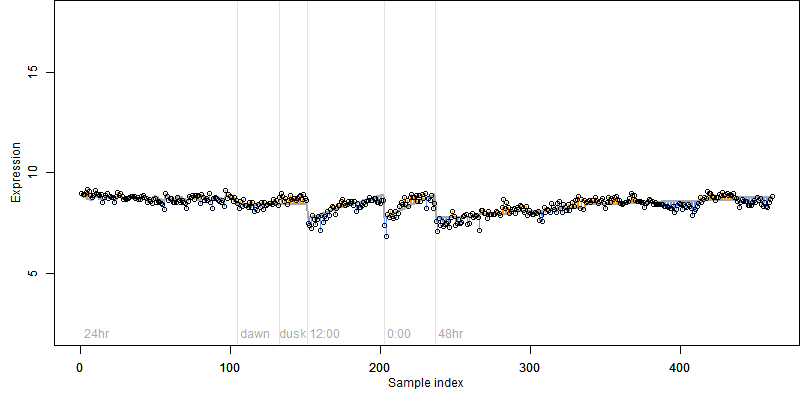
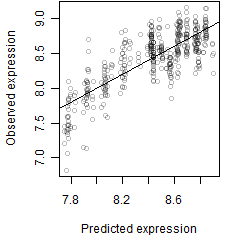
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
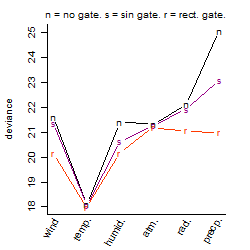
Dependence on each variable

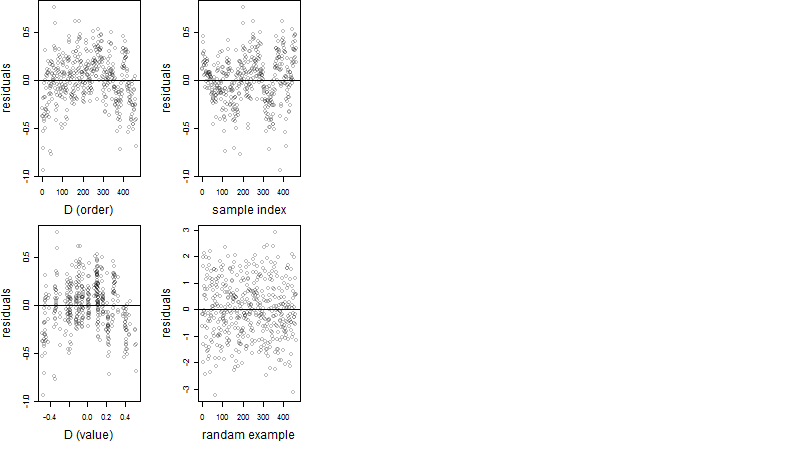
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17.81 | 0.198 | 8.34 | 0.924 | 0.0376 | 0.614 | 0.564 | 0.27 | 0.358 | 18 | 21.3 | 2582 | -- | -- |
| 17.92 | 0.197 | 8.35 | 0.881 | 0.0293 | 0.57 | 0.531 | -- | 0.347 | 17.7 | 21.5 | 2518 | -- | -- |
| 18.55 | 0.201 | 8.34 | 0.93 | 0.0335 | 0.607 | -- | 0.25 | 0.362 | 18.2 | 21.47 | 2426 | -- | -- |
| 18.66 | 0.201 | 8.35 | 0.867 | 0.0306 | 0.574 | -- | -- | 0.344 | 18.1 | 21.5 | 2420 | -- | -- |
| 18.59 | 0.201 | 8.34 | 0.934 | -- | 0.601 | -- | 0.247 | 0.359 | -- | 21.5 | 2390 | -- | -- |
| 27.29 | 0.245 | 8.31 | 1.16 | 0.0258 | -- | 0.607 | -- | 0.521 | 18.1 | -- | -- | -- | -- |
| 28.04 | 0.248 | 8.31 | 1.15 | 0.0565 | -- | -- | -- | 0.523 | 22.1 | -- | -- | -- | -- |
| 18.71 | 0.201 | 8.35 | 0.868 | -- | 0.574 | -- | -- | 0.344 | -- | 21.53 | 2397 | -- | -- |
| 22.68 | 0.222 | 8.39 | -- | 0.0481 | 1.06 | -- | -- | 0.165 | 21.2 | 17.84 | 47520 | -- | -- |
| 28.23 | 0.248 | 8.31 | 1.15 | -- | -- | -- | -- | 0.523 | -- | -- | -- | -- | -- |
| 62.85 | 0.371 | 8.34 | -- | 0.0652 | -- | -- | -- | 0.395 | 23.4 | -- | -- | -- | -- |
| 22.81 | 0.222 | 8.39 | -- | -- | 1.03 | -- | -- | 0.17 | -- | 17.9 | 47258 | -- | -- |
| 63.12 | 0.371 | 8.34 | -- | -- | -- | -- | -- | 0.395 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance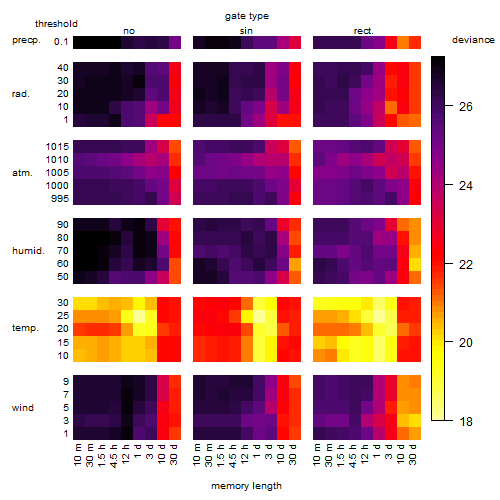
|
Histogram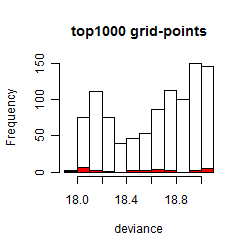 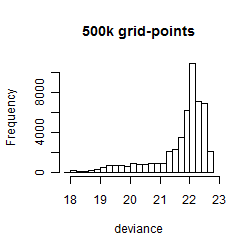
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 17.99 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 19 |
| 15 | 18.05 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 13 |
| 22 | 18.06 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 11 |
| 29 | 18.07 | temperature | 25 | 1440 | < th | dose dependent | sin | 13 | NA |
| 31 | 18.07 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 9 |
| 40 | 18.07 | temperature | 25 | 1440 | < th | dose dependent | rect. | 9 | 21 |
| 78 | 18.10 | temperature | 25 | 1440 | < th | dose dependent | rect. | 24 | 23 |
| 79 | 18.11 | temperature | 25 | 1440 | < th | dose dependent | no | NA | NA |
| 118 | 18.13 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 23 |
| 178 | 18.19 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 191 | 18.21 | temperature | 25 | 1440 | < th | dose dependent | rect. | 24 | 12 |
| 327 | 18.45 | temperature | 25 | 1440 | < th | dose dependent | rect. | 24 | 1 |
| 340 | 18.48 | temperature | 30 | 1440 | < th | dose dependent | rect. | 5 | 1 |
| 365 | 18.55 | temperature | 10 | 1440 | > th | dose dependent | rect. | 5 | 1 |
| 371 | 18.56 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 5 |
| 374 | 18.56 | temperature | 10 | 1440 | > th | dose dependent | rect. | 24 | 7 |
| 409 | 18.62 | temperature | 10 | 1440 | > th | dose dependent | rect. | 24 | 1 |
| 445 | 18.67 | temperature | 10 | 1440 | > th | dose dependent | rect. | 3 | 3 |
| 459 | 18.68 | temperature | 20 | 4320 | < th | dose independent | rect. | 17 | 7 |
| 467 | 18.69 | temperature | 20 | 4320 | > th | dose independent | rect. | 17 | 7 |
| 519 | 18.74 | temperature | 20 | 4320 | > th | dose independent | sin | 17 | NA |
| 523 | 18.74 | temperature | 20 | 4320 | < th | dose independent | rect. | 12 | 12 |
| 573 | 18.79 | temperature | 20 | 4320 | < th | dose independent | sin | 17 | NA |
| 777 | 18.96 | temperature | 20 | 4320 | < th | dose independent | rect. | 9 | 18 |
| 807 | 18.98 | temperature | 10 | 1440 | > th | dose dependent | sin | 10 | NA |
| 876 | 19.02 | temperature | 20 | 1440 | > th | dose independent | rect. | 3 | 20 |
| 945 | 19.06 | temperature | 20 | 1440 | < th | dose independent | rect. | 3 | 10 |
| 947 | 19.06 | temperature | 20 | 1440 | < th | dose independent | rect. | 3 | 20 |
| 958 | 19.07 | temperature | 20 | 1440 | > th | dose independent | rect. | 3 | 10 |
| 960 | 19.07 | temperature | 20 | 1440 | > th | dose independent | rect. | 20 | 19 |