Os01g0807900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Dihydropyrimidinase (Dihydropyrimidine amidohydrolase) (EC 3.5.2.2).

FiT-DB / Search/ Help/ Sample detail
|
Os01g0807900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Dihydropyrimidinase (Dihydropyrimidine amidohydrolase) (EC 3.5.2.2).
|
|
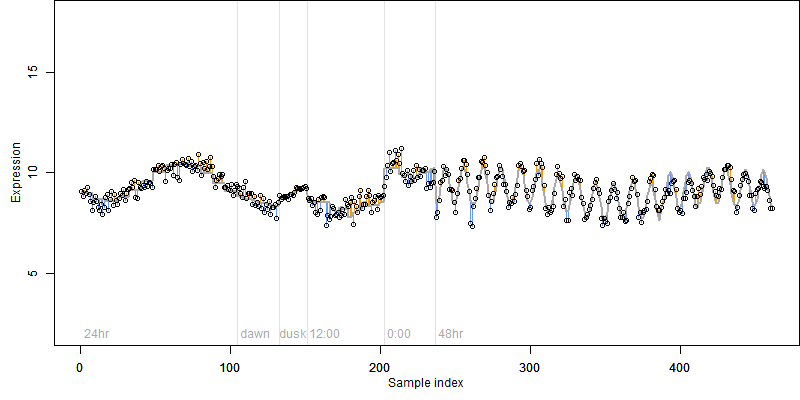
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
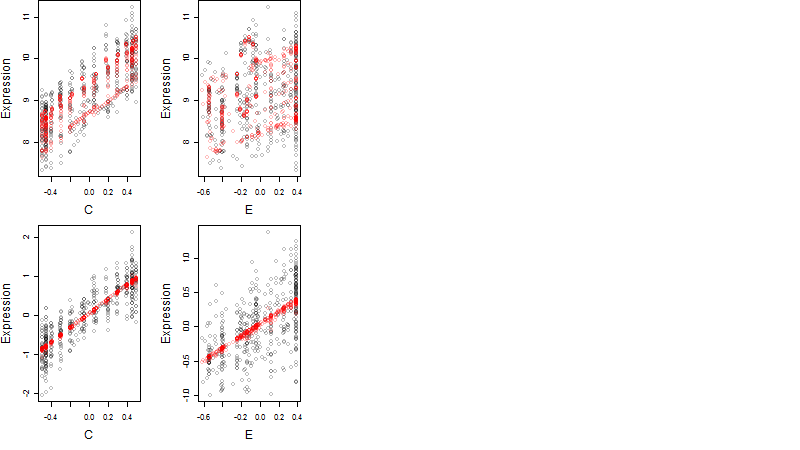
Dependence on each variable
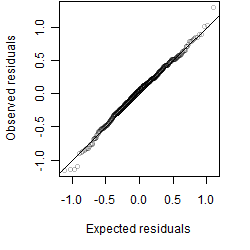
Residual plot

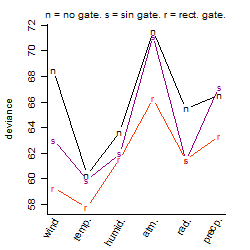
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 63.56 | 0.375 | 9.1 | -0.18 | 1.8 | 0.859 | -1.3 | -0.247 | 0.419 | 22.6 | 29.65 | 1698 | -- | -- |
| 63.58 | 0.371 | 9.1 | -0.21 | 1.78 | 0.835 | -1.28 | -- | 0.409 | 22.7 | 29.6 | 1693 | -- | -- |
| 69.74 | 0.389 | 9.09 | -0.238 | 1.9 | 0.907 | -- | 0.406 | 0.48 | 22.5 | 29.28 | 2047 | -- | -- |
| 69.76 | 0.389 | 9.08 | -0.17 | 1.89 | 0.958 | -- | -- | 0.506 | 22.5 | 29.21 | 2039 | -- | -- |
| 241.38 | 0.728 | 8.99 | -0.0388 | -- | 0.719 | -- | -0.714 | 0.689 | -- | 26.5 | 11701 | -- | -- |
| 88.68 | 0.441 | 9.1 | -0.465 | 1.75 | -- | -1.36 | -- | 0.429 | 22.7 | -- | -- | -- | -- |
| 96.45 | 0.46 | 9.1 | -0.41 | 1.76 | -- | -- | -- | 0.434 | 22.7 | -- | -- | -- | -- |
| 241.03 | 0.726 | 9.03 | -0.202 | -- | 0.719 | -- | -- | 0.548 | -- | 26.9 | 11690 | -- | -- |
| 69.36 | 0.388 | 9.03 | -- | 1.83 | 0.895 | -- | -- | 0.631 | 22.4 | 28.09 | 6198 | -- | -- |
| 288.3 | 0.793 | 9.05 | -0.374 | -- | -- | -- | -- | 0.444 | -- | -- | -- | -- | -- |
| 100.87 | 0.47 | 9.09 | -- | 1.76 | -- | -- | -- | 0.479 | 22.7 | -- | -- | -- | -- |
| 242.1 | 0.727 | 9.02 | -- | -- | 0.733 | -- | -- | 0.572 | -- | 26.9 | 11690 | -- | -- |
| 292 | 0.798 | 9.04 | -- | -- | -- | -- | -- | 0.485 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance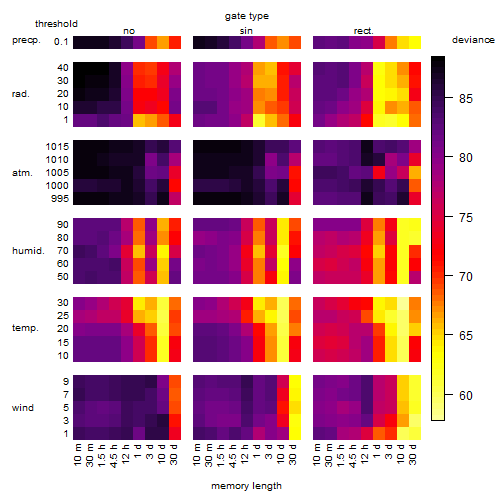
|
Histogram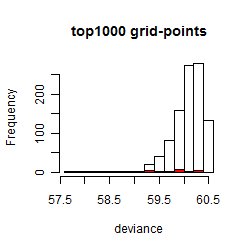 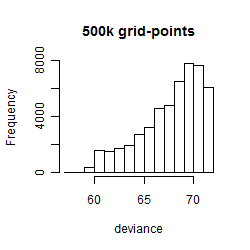
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 57.78 | temperature | 30 | 14400 | < th | dose independent | rect. | 14 | 2 |
| 4 | 57.98 | temperature | 30 | 14400 | > th | dose independent | rect. | 15 | 1 |
| 15 | 59.24 | temperature | 25 | 14400 | > th | dose dependent | rect. | 12 | 1 |
| 16 | 59.26 | temperature | 30 | 14400 | < th | dose independent | rect. | 14 | 15 |
| 31 | 59.28 | wind | 3 | 43200 | > th | dose dependent | rect. | 5 | 2 |
| 34 | 59.39 | temperature | 25 | 14400 | > th | dose independent | rect. | 4 | 9 |
| 44 | 59.48 | temperature | 25 | 14400 | < th | dose independent | rect. | 4 | 9 |
| 65 | 59.54 | temperature | 25 | 14400 | > th | dose dependent | rect. | 12 | 6 |
| 68 | 59.57 | temperature | 25 | 14400 | < th | dose independent | rect. | 7 | 4 |
| 142 | 59.76 | temperature | 25 | 14400 | > th | dose dependent | rect. | 15 | 1 |
| 146 | 59.78 | temperature | 25 | 14400 | > th | dose independent | rect. | 10 | 1 |
| 187 | 59.87 | temperature | 25 | 14400 | < th | dose independent | rect. | 10 | 1 |
| 203 | 59.88 | temperature | 25 | 14400 | > th | dose independent | sin | 3 | NA |
| 209 | 59.89 | temperature | 25 | 14400 | < th | dose independent | rect. | 8 | 1 |
| 229 | 59.93 | wind | 1 | 14400 | < th | dose dependent | rect. | 4 | 1 |
| 281 | 59.97 | temperature | 25 | 14400 | > th | dose dependent | sin | 21 | NA |
| 302 | 60.00 | temperature | 25 | 14400 | < th | dose independent | sin | 3 | NA |
| 313 | 60.00 | temperature | 25 | 14400 | > th | dose dependent | rect. | 11 | 18 |
| 329 | 60.02 | temperature | 25 | 14400 | > th | dose dependent | rect. | 3 | 15 |
| 355 | 60.05 | temperature | 25 | 14400 | > th | dose independent | rect. | 8 | 1 |
| 408 | 60.08 | temperature | 30 | 14400 | < th | dose independent | sin | 15 | NA |
| 589 | 60.21 | temperature | 30 | 14400 | > th | dose independent | sin | 15 | NA |
| 650 | 60.24 | temperature | 20 | 14400 | > th | dose dependent | rect. | 9 | 1 |
| 723 | 60.29 | temperature | 25 | 14400 | > th | dose dependent | no | NA | NA |
| 782 | 60.32 | temperature | 25 | 14400 | > th | dose dependent | rect. | 17 | 23 |
| 835 | 60.37 | temperature | 25 | 14400 | > th | dose dependent | rect. | 15 | 23 |