Os01g0637600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Peptide deformylase, chloroplast precursor (EC 3.5.1.88) (PDF) (Polypeptide deformylase).
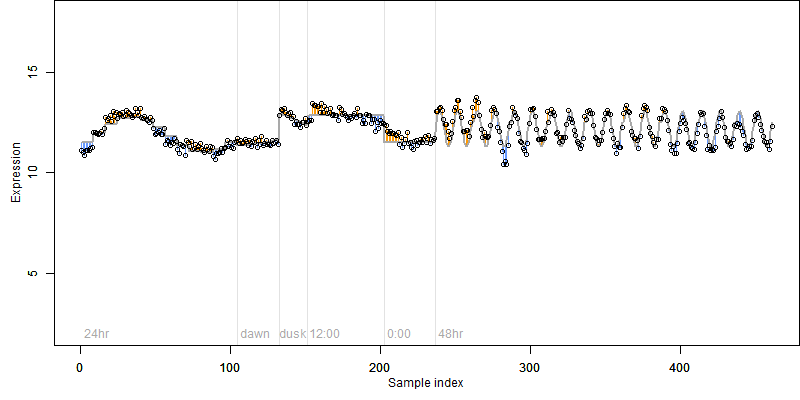

FiT-DB / Search/ Help/ Sample detail
|
Os01g0637600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Peptide deformylase, chloroplast precursor (EC 3.5.1.88) (PDF) (Polypeptide deformylase).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 28.22 | 0.25 | 12.2 | -0.644 | 0.975 | -0.9 | 0.0433 | 0.484 | -0.441 | 15.4 | 22.16 | 230 | -- | -- |
| 28.35 | 0.248 | 12.2 | -0.638 | 0.912 | -0.915 | -0.358 | -- | -0.432 | 15.5 | 18.01 | 227 | -- | -- |
| 27.91 | 0.246 | 12.2 | -0.621 | 0.826 | -0.991 | -- | 0.444 | -0.428 | 15.7 | 15.51 | 230 | -- | -- |
| 28.73 | 0.25 | 12.2 | -0.638 | 0.835 | -0.968 | -- | -- | -0.429 | 15.7 | 15.11 | 229 | -- | -- |
| 32.38 | 0.265 | 12.2 | -0.615 | -- | -1.58 | -- | 0.337 | -0.44 | -- | 8.597 | 320 | -- | -- |
| 39.18 | 0.293 | 12.2 | -0.572 | 1.78 | -- | -0.293 | -- | -0.375 | 14.7 | -- | -- | -- | -- |
| 39.48 | 0.294 | 12.2 | -0.58 | 1.78 | -- | -- | -- | -0.377 | 14.7 | -- | -- | -- | -- |
| 32.88 | 0.267 | 12.2 | -0.612 | -- | -1.58 | -- | -- | -0.441 | -- | 8.833 | 324 | -- | -- |
| 38.91 | 0.291 | 12.2 | -- | 0.828 | -0.93 | -- | -- | -0.343 | 15.9 | 10.03 | 234 | -- | -- |
| 223.21 | 0.698 | 12.2 | -0.655 | -- | -- | -- | -- | -0.422 | -- | -- | -- | -- | -- |
| 48.33 | 0.325 | 12.2 | -- | 1.79 | -- | -- | -- | -0.312 | 14.7 | -- | -- | -- | -- |
| 42.51 | 0.304 | 12.2 | -- | -- | -1.62 | -- | -- | -0.394 | -- | 10 | 326 | -- | -- |
| 234.54 | 0.715 | 12.2 | -- | -- | -- | -- | -- | -0.349 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 26.67 | radiation | 20 | 270 | < th | dose dependent | rect. | 3 | 16 |
| 5 | 26.87 | radiation | 20 | 270 | < th | dose dependent | rect. | 1 | 21 |
| 6 | 26.93 | radiation | 20 | 270 | < th | dose dependent | rect. | 24 | 23 |
| 12 | 27.09 | radiation | 10 | 270 | < th | dose dependent | sin | 1 | NA |
| 41 | 27.75 | humidity | 60 | 90 | > th | dose dependent | rect. | 4 | 18 |
| 44 | 27.78 | radiation | 10 | 270 | < th | dose independent | rect. | 3 | 17 |
| 52 | 27.83 | radiation | 20 | 270 | < th | dose dependent | rect. | 9 | 22 |
| 69 | 28.01 | radiation | 20 | 270 | < th | dose dependent | rect. | 12 | 21 |
| 101 | 28.26 | humidity | 50 | 270 | > th | dose dependent | rect. | 2 | 18 |
| 121 | 28.40 | radiation | 1 | 270 | > th | dose independent | sin | 1 | NA |
| 130 | 28.46 | radiation | 1 | 270 | < th | dose independent | sin | 1 | NA |
| 137 | 28.50 | humidity | 70 | 90 | > th | dose independent | rect. | 4 | 18 |
| 143 | 28.52 | temperature | 10 | 90 | > th | dose dependent | rect. | 8 | 10 |
| 173 | 28.65 | humidity | 50 | 270 | > th | dose dependent | rect. | 1 | 20 |
| 186 | 28.68 | temperature | 30 | 90 | < th | dose dependent | rect. | 4 | 18 |
| 202 | 28.75 | radiation | 10 | 270 | > th | dose independent | rect. | 16 | 23 |
| 210 | 28.79 | humidity | 90 | 270 | < th | dose independent | rect. | 7 | 9 |
| 227 | 28.86 | radiation | 10 | 270 | < th | dose independent | rect. | 12 | 21 |
| 231 | 28.87 | radiation | 10 | 270 | < th | dose independent | rect. | 13 | 18 |
| 232 | 28.87 | radiation | 20 | 270 | < th | dose dependent | no | NA | NA |
| 348 | 29.22 | radiation | 10 | 270 | > th | dose independent | rect. | 6 | 10 |
| 389 | 29.31 | radiation | 10 | 270 | < th | dose independent | rect. | 9 | 22 |
| 416 | 29.41 | radiation | 10 | 270 | > th | dose independent | no | NA | NA |
| 561 | 29.50 | radiation | 40 | 270 | < th | dose independent | rect. | 11 | 10 |
| 733 | 29.91 | radiation | 40 | 270 | < th | dose dependent | rect. | 10 | 10 |
| 743 | 29.93 | humidity | 70 | 90 | > th | dose dependent | sin | 0 | NA |
| 828 | 30.12 | temperature | 10 | 90 | > th | dose dependent | rect. | 8 | 20 |
| 914 | 30.26 | wind | 7 | 270 | < th | dose independent | rect. | 7 | 10 |
| 969 | 30.35 | temperature | 15 | 270 | > th | dose independent | rect. | 7 | 10 |