Os12g0533800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Mitochondrial substrate carrier family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os12g0533800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Mitochondrial substrate carrier family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
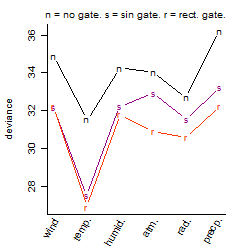
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 30.81 | 0.261 | 10 | -0.762 | 0.25 | 0.559 | 1.07 | -0.0461 | 0.0741 | 4.32 | 19.3 | 734 | -- | -- |
| 30.82 | 0.259 | 10 | -0.767 | 0.248 | 0.567 | 1.07 | -- | 0.073 | 4.29 | 19.33 | 733 | -- | -- |
| 33.37 | 0.269 | 9.99 | -0.685 | 0.306 | 0.592 | -- | -0.0626 | 0.101 | 5.25 | 19.5 | 788 | -- | -- |
| 32.97 | 0.267 | 10 | -0.785 | 0.304 | 0.518 | -- | -- | 0.0624 | 5.98 | 19.4 | 788 | -- | -- |
| 37.45 | 0.285 | 10 | -0.744 | -- | 0.559 | -- | -0.105 | 0.0857 | -- | 19.5 | 693 | -- | -- |
| 37.29 | 0.286 | 10 | -0.975 | 0.286 | -- | 0.926 | -- | -0.0243 | 5.02 | -- | -- | -- | -- |
| 38.25 | 0.29 | 10 | -0.974 | 0.33 | -- | -- | -- | -0.0279 | 6.7 | -- | -- | -- | -- |
| 37.46 | 0.285 | 10 | -0.771 | -- | 0.548 | -- | -- | 0.0734 | -- | 19.45 | 690 | -- | -- |
| 45.43 | 0.314 | 9.97 | -- | 0.322 | 0.871 | -- | -- | 0.211 | 6.21 | 18.98 | 2428 | -- | -- |
| 43.95 | 0.31 | 10 | -0.966 | -- | -- | -- | -- | -0.0214 | -- | -- | -- | -- | -- |
| 63.25 | 0.372 | 9.99 | -- | 0.319 | -- | -- | -- | 0.0806 | 6.88 | -- | -- | -- | -- |
| 46.53 | 0.318 | 9.96 | -- | -- | 0.802 | -- | -- | 0.246 | -- | 18.57 | 21126 | -- | -- |
| 68.58 | 0.387 | 10 | -- | -- | -- | -- | -- | 0.0859 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 26.88 | temperature | 20 | 720 | < th | dose independent | rect. | 24 | 7 |
| 2 | 26.88 | temperature | 20 | 720 | < th | dose independent | rect. | 23 | 9 |
| 17 | 27.18 | temperature | 30 | 90 | < th | dose dependent | rect. | 4 | 11 |
| 22 | 27.23 | temperature | 30 | 270 | < th | dose dependent | rect. | 2 | 12 |
| 29 | 27.30 | temperature | 30 | 90 | < th | dose dependent | rect. | 4 | 9 |
| 51 | 27.49 | temperature | 20 | 720 | < th | dose independent | sin | 7 | NA |
| 73 | 27.61 | temperature | 20 | 720 | < th | dose dependent | rect. | 3 | 2 |
| 83 | 27.71 | temperature | 20 | 720 | < th | dose dependent | rect. | 1 | 4 |
| 98 | 27.83 | temperature | 25 | 720 | < th | dose dependent | rect. | 2 | 6 |
| 110 | 27.90 | temperature | 30 | 90 | < th | dose dependent | sin | 3 | NA |
| 152 | 28.10 | temperature | 20 | 720 | < th | dose independent | rect. | 5 | 1 |
| 281 | 28.60 | temperature | 20 | 720 | > th | dose independent | rect. | 5 | 6 |
| 320 | 28.74 | temperature | 20 | 720 | > th | dose independent | rect. | 24 | 11 |
| 333 | 28.80 | temperature | 20 | 720 | > th | dose independent | rect. | 24 | 18 |
| 394 | 28.96 | temperature | 20 | 720 | > th | dose independent | sin | 4 | NA |
| 502 | 29.27 | temperature | 25 | 720 | < th | dose dependent | rect. | 7 | 1 |
| 566 | 29.43 | temperature | 20 | 720 | > th | dose independent | rect. | 23 | 4 |
| 608 | 29.53 | temperature | 20 | 270 | > th | dose dependent | rect. | 5 | 7 |
| 683 | 29.71 | temperature | 10 | 90 | > th | dose dependent | sin | 2 | NA |
| 780 | 29.93 | temperature | 10 | 720 | > th | dose dependent | rect. | 24 | 10 |
| 801 | 29.97 | temperature | 10 | 720 | > th | dose dependent | rect. | 5 | 6 |