Os12g0510900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to LIM domain protein WLIM-1.
FiT-DB / Search/ Help/ Sample detail
|
Os12g0510900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to LIM domain protein WLIM-1.
|
|
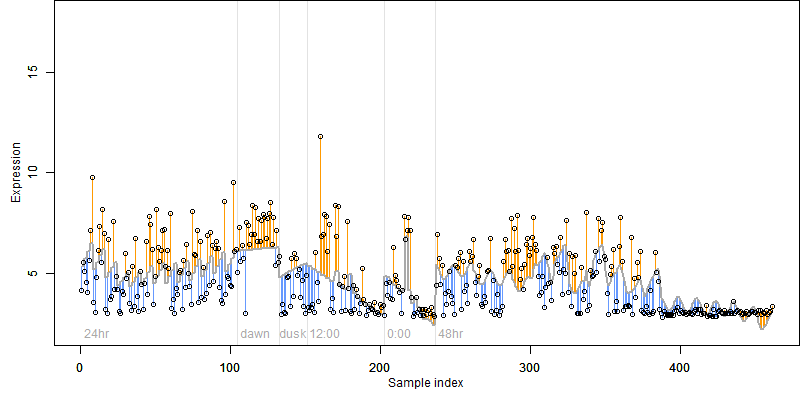
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

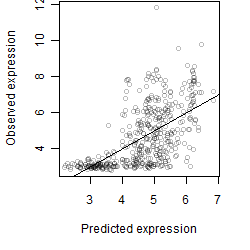
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 794.05 | 1.32 | 4.92 | -6.13 | 0.872 | 3.89 | -0.904 | -14.7 | -0.231 | 8.99 | 26.26 | 1154 | 11.4 | -- |
| 832.07 | 1.34 | 4.58 | -2.93 | 0.751 | 2.96 | -0.472 | -- | 0.345 | 10.1 | 25.4 | 952 | 7.23 | -- |
| 789.29 | 1.31 | 4.84 | -5.72 | 0.743 | 3.93 | -- | -14.4 | 0.0641 | 8.76 | 26.48 | 1127 | 8.47 | -- |
| 830.74 | 1.34 | 4.52 | -2.88 | 0.817 | 2.68 | -- | -- | 0.465 | 9.43 | 25.9 | 972 | 7.48 | -- |
| 817.35 | 1.33 | 4.85 | -5.66 | -- | 3.91 | -- | -14.1 | 0.0895 | -- | 26.38 | 1162 | 7.99 | -- |
| 1046.01 | 1.52 | 4.52 | -2.28 | 0.907 | -- | 1.68 | -- | 0.454 | 5.37 | -- | -- | -- | -- |
| 1051.41 | 1.52 | 4.52 | -2.27 | 0.937 | -- | -- | -- | 0.448 | 6.03 | -- | -- | -- | -- |
| 862.36 | 1.37 | 4.52 | -2.84 | -- | 2.47 | -- | -- | 0.532 | -- | 26.2 | 1100 | 7.59 | -- |
| 1038.6 | 1.5 | 4.44 | -- | 0.834 | 2.1 | -- | -- | 0.82 | 8.6 | 26.26 | 969 | 7.18 | -- |
| 1096.91 | 1.55 | 4.53 | -2.25 | -- | -- | -- | -- | 0.47 | -- | -- | -- | -- | -- |
| 1187.6 | 1.61 | 4.46 | -- | 0.904 | -- | -- | -- | 0.701 | 6.16 | -- | -- | -- | -- |
| 1070.56 | 1.52 | 4.45 | -- | -- | 2.03 | -- | -- | 0.839 | -- | 26.3 | 1061 | 7.08 | -- |
| 1229.98 | 1.64 | 4.48 | -- | -- | -- | -- | -- | 0.719 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance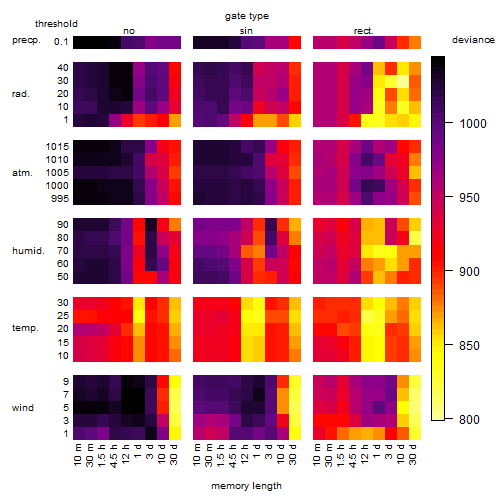
|
Histogram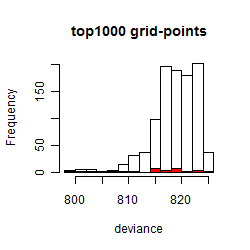 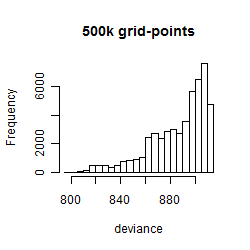
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 798.47 | radiation | 30 | 14400 | < th | dose independent | rect. | 16 | 2 |
| 2 | 799.27 | radiation | 30 | 14400 | < th | dose independent | rect. | 16 | 4 |
| 5 | 800.57 | radiation | 30 | 14400 | < th | dose independent | rect. | 16 | 12 |
| 10 | 802.47 | radiation | 30 | 14400 | < th | dose independent | rect. | 16 | 9 |
| 15 | 806.91 | wind | 5 | 43200 | > th | dose dependent | rect. | 12 | 20 |
| 24 | 809.05 | wind | 5 | 43200 | > th | dose independent | rect. | 12 | 19 |
| 33 | 810.15 | wind | 5 | 43200 | < th | dose independent | rect. | 11 | 20 |
| 103 | 814.09 | wind | 5 | 43200 | > th | dose dependent | rect. | 24 | 20 |
| 105 | 814.16 | wind | 5 | 43200 | > th | dose dependent | rect. | 2 | 18 |
| 110 | 814.28 | wind | 5 | 43200 | > th | dose dependent | sin | 16 | NA |
| 135 | 814.97 | wind | 5 | 43200 | < th | dose independent | rect. | 23 | 21 |
| 137 | 815.04 | wind | 5 | 43200 | > th | dose independent | rect. | 23 | 21 |
| 142 | 815.09 | wind | 5 | 43200 | > th | dose independent | sin | 15 | NA |
| 245 | 816.78 | wind | 5 | 43200 | < th | dose independent | sin | 15 | NA |
| 255 | 816.87 | wind | 5 | 43200 | > th | dose dependent | rect. | 6 | 14 |
| 336 | 817.60 | wind | 5 | 43200 | > th | dose dependent | no | NA | NA |
| 389 | 817.98 | temperature | 25 | 720 | > th | dose dependent | rect. | 18 | 5 |
| 409 | 818.14 | wind | 5 | 43200 | < th | dose independent | no | NA | NA |
| 457 | 818.63 | wind | 7 | 43200 | < th | dose independent | rect. | 18 | 23 |
| 473 | 818.78 | wind | 5 | 43200 | > th | dose independent | no | NA | NA |
| 483 | 818.88 | wind | 5 | 43200 | > th | dose independent | sin | 23 | NA |
| 494 | 818.97 | wind | 5 | 43200 | > th | dose dependent | rect. | 18 | 23 |
| 556 | 819.67 | wind | 5 | 43200 | < th | dose independent | sin | 23 | NA |
| 580 | 819.97 | wind | 7 | 43200 | > th | dose independent | rect. | 18 | 23 |
| 677 | 821.20 | humidity | 80 | 43200 | < th | dose independent | rect. | 21 | 2 |
| 855 | 822.85 | wind | 5 | 43200 | > th | dose dependent | rect. | 15 | 1 |
| 865 | 823.00 | wind | 3 | 43200 | > th | dose dependent | rect. | 9 | 7 |
| 915 | 823.50 | humidity | 80 | 43200 | > th | dose independent | rect. | 22 | 1 |
| 988 | 824.29 | wind | 5 | 43200 | > th | dose independent | rect. | 9 | 7 |