Os12g0120500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF567 family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os12g0120500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF567 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
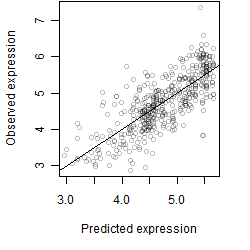
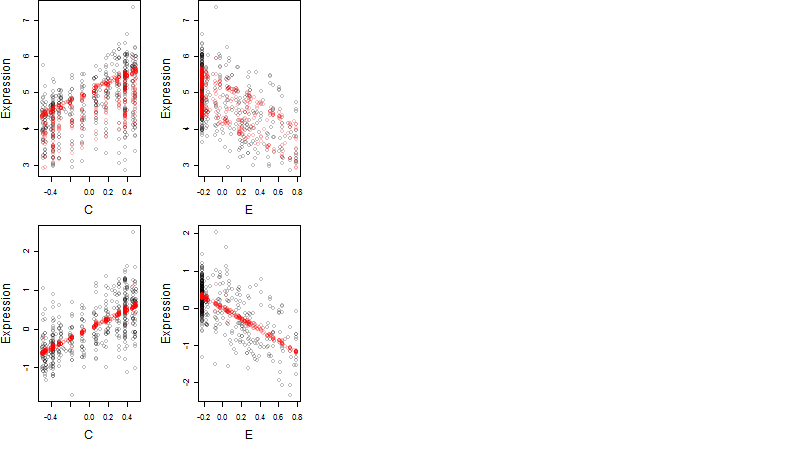
Dependence on each variable
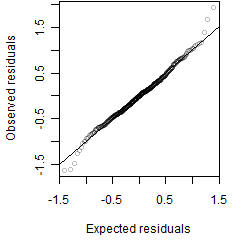
Residual plot
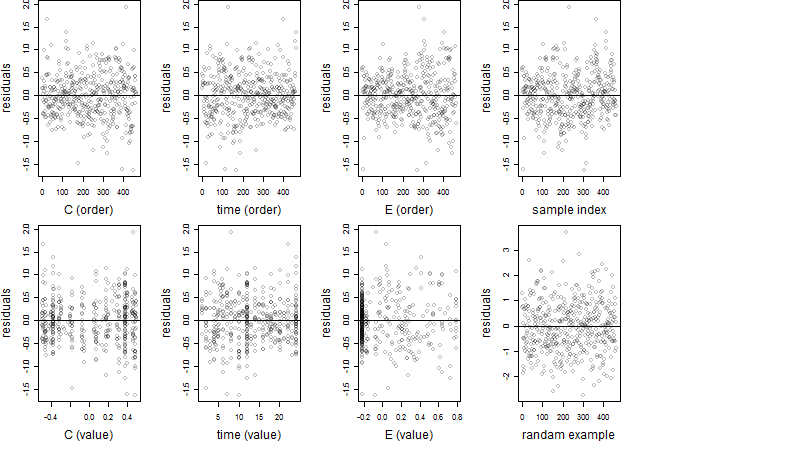
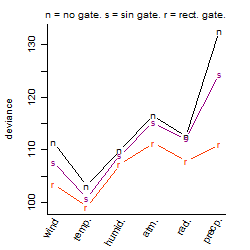
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100.29 | 0.471 | 4.71 | 0.815 | 1.19 | -1.01 | 0.287 | 0.192 | 0.0599 | 9.29 | 22.91 | 1776 | -- | -- |
| 99.07 | 0.464 | 4.7 | 0.816 | 1.23 | -1.04 | 0.439 | -- | 0.109 | 9.22 | 22.29 | 1897 | -- | -- |
| 99.5 | 0.465 | 4.7 | 0.843 | 1.21 | -1.08 | -- | 0.0687 | 0.0847 | 9.3 | 22.4 | 1897 | -- | -- |
| 99.19 | 0.464 | 4.7 | 0.85 | 1.22 | -1.1 | -- | -- | 0.095 | 9.29 | 22.24 | 1931 | -- | -- |
| 173.11 | 0.616 | 4.69 | 1.13 | -- | -0.863 | -- | -0.433 | 0.252 | -- | 22.39 | 1583 | -- | -- |
| 143.24 | 0.561 | 4.61 | 1.27 | 1.17 | -- | 0.34 | -- | 0.476 | 9.26 | -- | -- | -- | -- |
| 143.66 | 0.561 | 4.61 | 1.29 | 1.17 | -- | -- | -- | 0.476 | 9.26 | -- | -- | -- | -- |
| 174.27 | 0.618 | 4.7 | 1.01 | -- | -0.862 | -- | -- | 0.221 | -- | 22.39 | 1583 | -- | -- |
| 111.03 | 0.491 | 4.74 | -- | 1.23 | -1.48 | -- | -- | -0.079 | 9.41 | 21.5 | 3868 | -- | -- |
| 223.38 | 0.698 | 4.64 | 1.28 | -- | -- | -- | -- | 0.481 | -- | -- | -- | -- | -- |
| 187.28 | 0.64 | 4.64 | -- | 1.17 | -- | -- | -- | 0.333 | 9.18 | -- | -- | -- | -- |
| 191.89 | 0.645 | 4.76 | -- | -- | -1.53 | -- | -- | -0.0346 | -- | 20.6 | 5931 | -- | -- |
| 266.71 | 0.762 | 4.67 | -- | -- | -- | -- | -- | 0.339 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance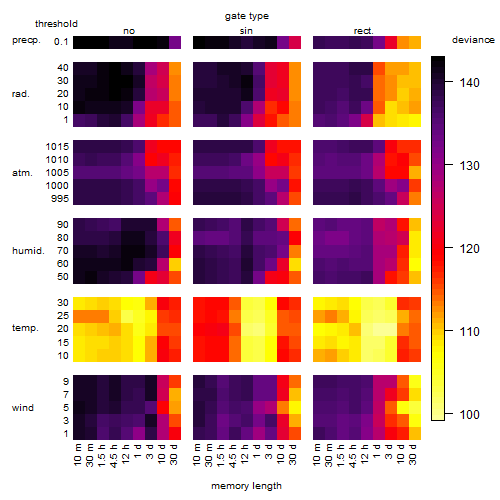
|
Histogram 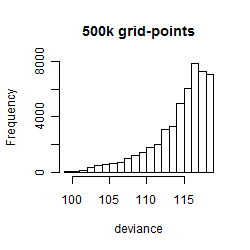
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 99.09 | temperature | 20 | 4320 | > th | dose independent | rect. | 21 | 2 |
| 2 | 99.32 | temperature | 20 | 1440 | > th | dose dependent | rect. | 21 | 9 |
| 21 | 99.92 | temperature | 20 | 4320 | < th | dose independent | rect. | 21 | 2 |
| 47 | 100.40 | temperature | 20 | 1440 | > th | dose dependent | rect. | 21 | 1 |
| 62 | 100.69 | temperature | 20 | 1440 | > th | dose dependent | rect. | 5 | 1 |
| 73 | 100.83 | temperature | 20 | 1440 | > th | dose dependent | sin | 12 | NA |
| 96 | 101.15 | temperature | 20 | 720 | > th | dose dependent | sin | 9 | NA |
| 113 | 101.38 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 119 | 101.51 | temperature | 10 | 720 | > th | dose dependent | rect. | 18 | 14 |
| 129 | 101.66 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 12 |
| 133 | 101.70 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 6 |
| 134 | 101.71 | temperature | 25 | 1440 | < th | dose dependent | rect. | 18 | 9 |
| 171 | 101.94 | temperature | 25 | 720 | < th | dose dependent | rect. | 11 | 21 |
| 195 | 102.08 | temperature | 25 | 1440 | < th | dose dependent | sin | 14 | NA |
| 200 | 102.11 | temperature | 25 | 720 | < th | dose dependent | rect. | 13 | 19 |
| 398 | 102.72 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 1 |
| 458 | 102.92 | temperature | 25 | 720 | < th | dose dependent | rect. | 17 | 22 |
| 485 | 102.97 | temperature | 25 | 720 | < th | dose dependent | rect. | 4 | 23 |
| 556 | 103.13 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 631 | 103.33 | temperature | 25 | 720 | < th | dose dependent | rect. | 1 | 23 |
| 639 | 103.35 | temperature | 25 | 720 | < th | dose dependent | rect. | 6 | 23 |
| 694 | 103.51 | wind | 5 | 43200 | > th | dose independent | rect. | 13 | 1 |
| 750 | 103.61 | wind | 5 | 43200 | < th | dose independent | rect. | 13 | 1 |
| 953 | 104.00 | temperature | 20 | 4320 | > th | dose independent | sin | 15 | NA |