Os11g0672900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF1645 family protein.
FiT-DB / Search/ Help/ Sample detail
|
Os11g0672900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF1645 family protein.
|
|
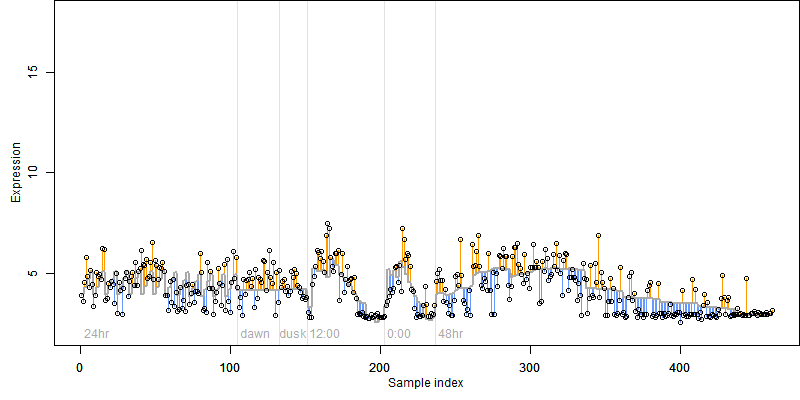
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
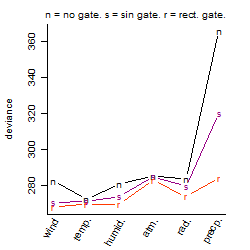
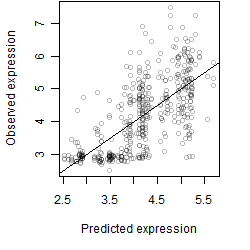
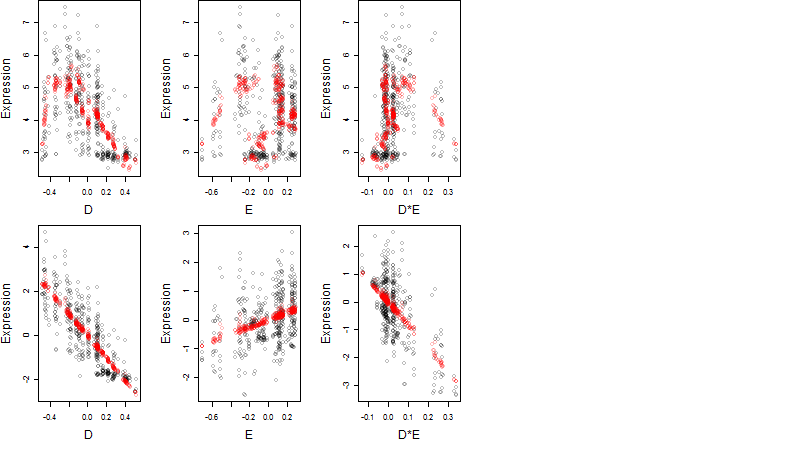
Dependence on each variable
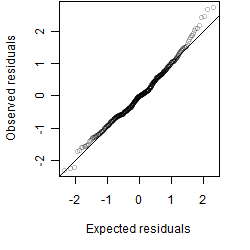
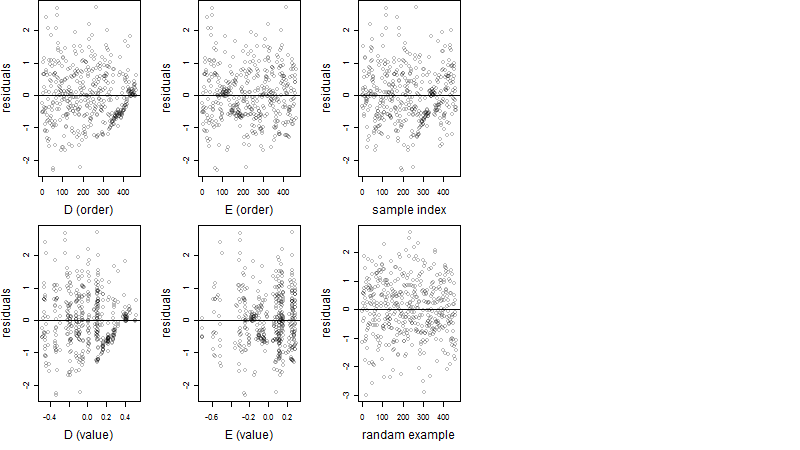
Residual plot
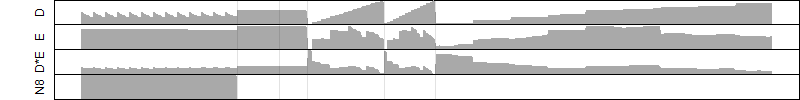
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 284.92 | 0.793 | 4.57 | -4.94 | 0.387 | 1.27 | -2.24 | -8.5 | -0.761 | 15.2 | 7.012 | 5514 | -- | -- |
| 344.83 | 0.865 | 4.28 | -3.2 | 0.389 | 1.81 | -1.88 | -- | -0.392 | 15 | -7.667 | 7941 | -- | -- |
| 298.56 | 0.805 | 4.57 | -4.94 | 0.347 | 1.26 | -- | -8.17 | -0.78 | 16.1 | 11.2 | 5976 | -- | -- |
| 356.52 | 0.879 | 4.29 | -3.27 | 0.43 | 1.81 | -- | -- | -0.428 | 15.7 | -7.869 | 8379 | -- | -- |
| 305 | 0.813 | 4.58 | -4.99 | -- | 1.23 | -- | -8.29 | -0.81 | -- | 11.28 | 6085 | -- | -- |
| 397.45 | 0.935 | 4.17 | -2.27 | 0.339 | -- | -1.97 | -- | 0.108 | 15.1 | -- | -- | -- | -- |
| 409.7 | 0.948 | 4.18 | -2.31 | 0.354 | -- | -- | -- | 0.0976 | 16.2 | -- | -- | -- | -- |
| 364.31 | 0.889 | 4.26 | -3.19 | -- | 1.75 | -- | -- | -0.315 | -- | -61.81 | 6137 | -- | -- |
| 547.64 | 1.09 | 4.15 | -- | 0.407 | 0.408 | -- | -- | 0.289 | 15.8 | -13.88 | 1235 | -- | -- |
| 416.49 | 0.954 | 4.17 | -2.33 | -- | -- | -- | -- | 0.0882 | -- | -- | -- | -- | -- |
| 550.84 | 1.1 | 4.12 | -- | 0.398 | -- | -- | -- | 0.355 | 16.1 | -- | -- | -- | -- |
| 553.72 | 1.1 | 4.13 | -- | -- | 0.613 | -- | -- | 0.277 | -- | -61.81 | 6137 | -- | -- |
| 559.48 | 1.1 | 4.12 | -- | -- | -- | -- | -- | 0.347 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance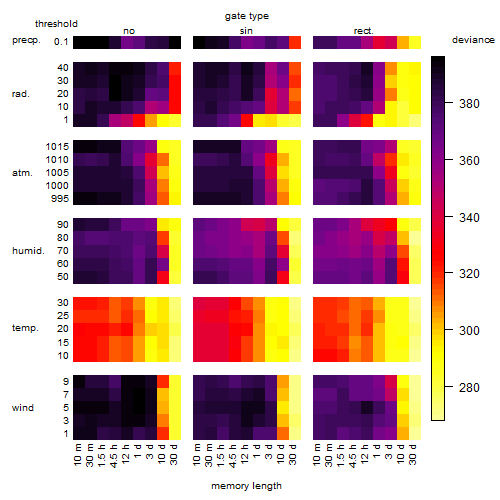
|
Histogram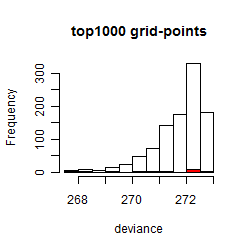 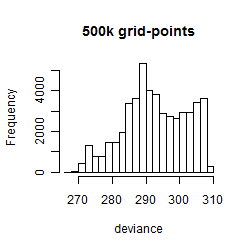
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 267.75 | wind | 7 | 43200 | < th | dose dependent | rect. | 13 | 14 |
| 7 | 268.03 | wind | 1 | 43200 | > th | dose dependent | rect. | 13 | 14 |
| 10 | 268.24 | wind | 1 | 43200 | > th | dose dependent | rect. | 14 | 12 |
| 26 | 269.33 | humidity | 80 | 43200 | > th | dose independent | rect. | 20 | 5 |
| 30 | 269.46 | temperature | 10 | 43200 | > th | dose dependent | rect. | 8 | 1 |
| 32 | 269.50 | temperature | 30 | 43200 | < th | dose dependent | rect. | 8 | 1 |
| 91 | 270.42 | wind | 7 | 43200 | < th | dose dependent | sin | 16 | NA |
| 146 | 270.82 | humidity | 80 | 43200 | > th | dose independent | rect. | 19 | 10 |
| 209 | 271.17 | temperature | 10 | 43200 | > th | dose dependent | sin | 5 | NA |
| 246 | 271.30 | wind | 1 | 43200 | > th | dose dependent | sin | 16 | NA |
| 306 | 271.48 | temperature | 30 | 43200 | < th | dose dependent | sin | 6 | NA |
| 563 | 272.13 | humidity | 80 | 43200 | < th | dose independent | rect. | 20 | 5 |
| 583 | 272.15 | temperature | 10 | 43200 | > th | dose dependent | no | NA | NA |
| 586 | 272.15 | temperature | 10 | 43200 | > th | dose dependent | rect. | 5 | 23 |
| 755 | 272.43 | temperature | 30 | 43200 | < th | dose dependent | rect. | 21 | 2 |
| 770 | 272.45 | temperature | 10 | 43200 | > th | dose dependent | rect. | 21 | 2 |
| 811 | 272.50 | temperature | 30 | 43200 | < th | dose dependent | no | NA | NA |
| 820 | 272.50 | temperature | 30 | 43200 | < th | dose dependent | rect. | 24 | 23 |
| 848 | 272.52 | temperature | 30 | 43200 | < th | dose dependent | rect. | 5 | 23 |