Os11g0643800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Major facilitator superfamily protein.


FiT-DB / Search/ Help/ Sample detail
|
Os11g0643800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Major facilitator superfamily protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
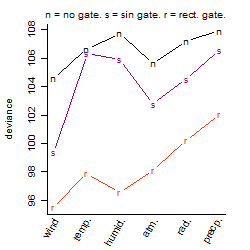
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 103.11 | 0.477 | 3.02 | 0.0364 | 0.184 | -0.319 | 0.0619 | 2.53 | 0.00749 | 10.6 | 7.722 | 147 | -- | -- |
| 105.84 | 0.479 | 3.03 | 0.0202 | 0.231 | -0.559 | 0.166 | -- | -0.0232 | 10.4 | 6.801 | 147 | -- | -- |
| 103.12 | 0.473 | 3.02 | 0.0391 | 0.184 | -0.329 | -- | 2.48 | 0.00737 | 10.6 | 7.724 | 146 | -- | -- |
| 104.46 | 0.476 | 3.05 | 0.053 | 0.245 | -0.855 | -- | -- | -0.0303 | 10.1 | 7.8 | 156 | -- | -- |
| 105.19 | 0.478 | 3.02 | 0.0411 | -- | -0.357 | -- | 2.55 | 0.00881 | -- | 7.704 | 146 | -- | -- |
| 109.39 | 0.49 | 3.05 | -0.046 | 0.214 | -- | -0.204 | -- | -0.0384 | 12 | -- | -- | -- | -- |
| 109.56 | 0.49 | 3.05 | -0.0546 | 0.215 | -- | -- | -- | -0.0397 | 11.7 | -- | -- | -- | -- |
| 106.21 | 0.48 | 3.04 | 0.071 | -- | -1.07 | -- | -- | 0.00278 | -- | 7.741 | 151 | -- | -- |
| 104.15 | 0.475 | 3.04 | -- | 0.188 | -0.993 | -- | -- | -0.00431 | 10.5 | 7.712 | 154 | -- | -- |
| 112.47 | 0.496 | 3.05 | -0.0608 | -- | -- | -- | -- | -0.0423 | -- | -- | -- | -- | -- |
| 109.63 | 0.49 | 3.05 | -- | 0.216 | -- | -- | -- | -0.0336 | 11.7 | -- | -- | -- | -- |
| 106.29 | 0.48 | 3.04 | -- | -- | -1.03 | -- | -- | -0.00361 | -- | 7.759 | 146 | -- | -- |
| 112.57 | 0.495 | 3.05 | -- | -- | -- | -- | -- | -0.0355 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 95.52 | wind | 5 | 270 | > th | dose dependent | rect. | 10 | 3 |
| 3 | 95.65 | wind | 3 | 270 | > th | dose dependent | rect. | 10 | 1 |
| 5 | 95.80 | wind | 5 | 270 | > th | dose independent | rect. | 10 | 1 |
| 11 | 96.12 | wind | 7 | 270 | > th | dose independent | rect. | 10 | 3 |
| 18 | 96.56 | humidity | 90 | 90 | > th | dose independent | rect. | 9 | 1 |
| 19 | 96.56 | humidity | 90 | 90 | > th | dose dependent | rect. | 9 | 1 |
| 24 | 96.81 | humidity | 80 | 10 | < th | dose independent | rect. | 13 | 1 |
| 43 | 97.50 | wind | 7 | 720 | < th | dose independent | rect. | 5 | 6 |
| 61 | 97.88 | temperature | 25 | 10 | > th | dose independent | rect. | 13 | 1 |
| 79 | 98.11 | atmosphere | 1010 | 30 | < th | dose dependent | rect. | 12 | 2 |
| 110 | 98.39 | wind | 7 | 90 | < th | dose independent | rect. | 14 | 22 |
| 124 | 98.49 | wind | 7 | 270 | > th | dose dependent | rect. | 24 | 13 |
| 131 | 98.54 | atmosphere | 1005 | 10 | < th | dose independent | rect. | 13 | 1 |
| 190 | 99.02 | temperature | 20 | 10 | > th | dose dependent | rect. | 13 | 1 |
| 232 | 99.35 | wind | 7 | 270 | < th | dose independent | sin | 6 | NA |
| 243 | 99.48 | wind | 7 | 270 | > th | dose independent | sin | 6 | NA |
| 273 | 99.77 | humidity | 90 | 10 | < th | dose dependent | rect. | 13 | 1 |
| 302 | 99.94 | wind | 7 | 270 | > th | dose dependent | sin | 6 | NA |
| 336 | 100.08 | wind | 5 | 90 | < th | dose independent | rect. | 5 | 7 |
| 357 | 100.18 | radiation | 10 | 10 | < th | dose dependent | rect. | 11 | 2 |
| 362 | 100.20 | radiation | 10 | 10 | < th | dose independent | rect. | 12 | 1 |
| 492 | 100.58 | wind | 7 | 270 | < th | dose independent | rect. | 5 | 12 |
| 499 | 100.60 | radiation | 10 | 10 | < th | dose independent | rect. | 10 | 2 |
| 574 | 100.75 | temperature | 30 | 10 | < th | dose independent | rect. | 12 | 2 |
| 592 | 100.77 | wind | 7 | 90 | < th | dose independent | rect. | 3 | 9 |
| 629 | 100.86 | wind | 9 | 720 | < th | dose dependent | rect. | 5 | 6 |
| 660 | 100.93 | wind | 7 | 720 | < th | dose independent | rect. | 21 | 14 |
| 668 | 100.95 | radiation | 1 | 10 | > th | dose independent | rect. | 12 | 2 |
| 713 | 101.03 | wind | 9 | 10 | > th | dose dependent | rect. | 11 | 3 |
| 802 | 101.10 | wind | 9 | 90 | < th | dose dependent | rect. | 5 | 7 |
| 861 | 101.24 | atmosphere | 1010 | 10 | > th | dose independent | rect. | 11 | 1 |