Os11g0582100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Zinc finger, RING-type domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os11g0582100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Zinc finger, RING-type domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
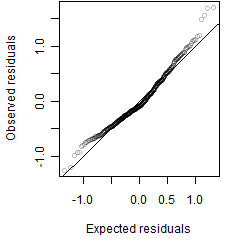
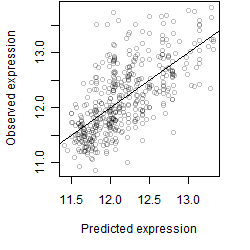 __
__

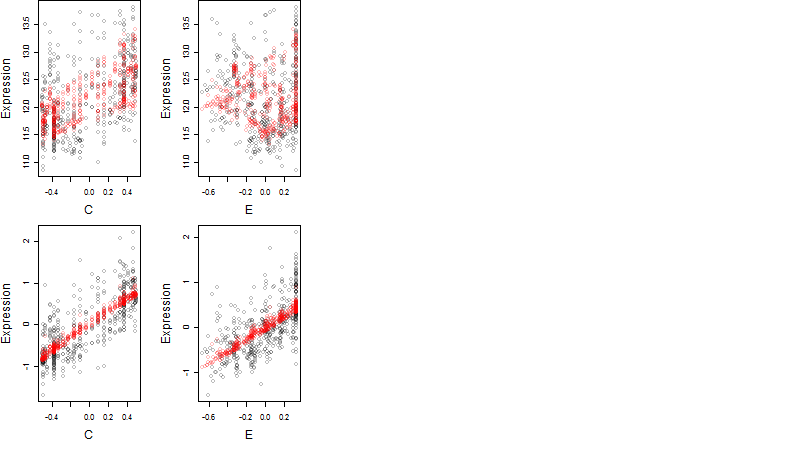
Dependence on each variable
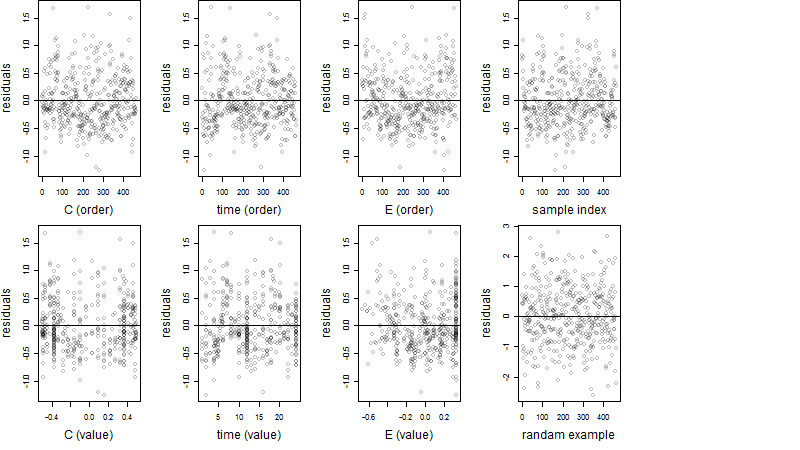
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 98.63 | 0.467 | 12.1 | 0.197 | 1.55 | 1.34 | 0.0408 | -0.463 | 0.36 | 21.3 | 73.99 | 835 | -- | -- |
| 98.81 | 0.463 | 12.1 | 0.167 | 1.57 | 1.36 | 0.241 | -- | 0.348 | 21.2 | 73.54 | 819 | -- | -- |
| 98.49 | 0.462 | 12.1 | 0.202 | 1.57 | 1.37 | -- | -0.542 | 0.358 | 21.2 | 73.97 | 818 | -- | -- |
| 99.02 | 0.463 | 12.1 | 0.158 | 1.57 | 1.35 | -- | -- | 0.347 | 21.2 | 73.53 | 819 | -- | -- |
| 143.2 | 0.56 | 12.1 | 0.0679 | -- | 0.57 | -- | -0.463 | 0.294 | -- | 73.97 | 818 | -- | -- |
| 132.54 | 0.54 | 12.1 | 0.00198 | 0.867 | -- | 0.0467 | -- | 0.278 | 21.7 | -- | -- | -- | -- |
| 132.55 | 0.539 | 12.1 | 0.000141 | 0.866 | -- | -- | -- | 0.278 | 21.7 | -- | -- | -- | -- |
| 144.44 | 0.562 | 12.1 | 0.0524 | -- | 0.567 | -- | -- | 0.289 | -- | 73.97 | 818 | -- | -- |
| 99.66 | 0.465 | 12.1 | -- | 1.55 | 1.33 | -- | -- | 0.328 | 21.2 | 73.37 | 819 | -- | -- |
| 177.17 | 0.622 | 12.1 | 0.0082 | -- | -- | -- | -- | 0.277 | -- | -- | -- | -- | -- |
| 132.55 | 0.539 | 12.1 | -- | 0.866 | -- | -- | -- | 0.278 | 21.7 | -- | -- | -- | -- |
| 144.52 | 0.562 | 12.1 | -- | -- | 0.566 | -- | -- | 0.283 | -- | 73.97 | 818 | -- | -- |
| 177.18 | 0.621 | 12.1 | -- | -- | -- | -- | -- | 0.276 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance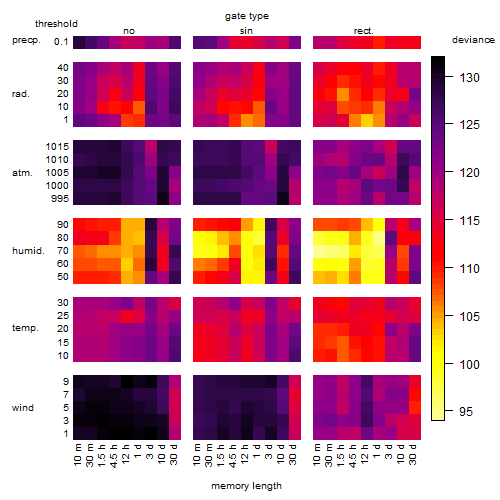
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 93.97 | humidity | 70 | 10 | > th | dose dependent | rect. | 6 | 14 |
| 2 | 94.61 | humidity | 80 | 1440 | < th | dose independent | rect. | 8 | 7 |
| 6 | 95.35 | humidity | 80 | 1440 | > th | dose independent | rect. | 8 | 7 |
| 26 | 96.47 | humidity | 70 | 1440 | > th | dose dependent | rect. | 9 | 5 |
| 30 | 96.63 | humidity | 70 | 90 | > th | dose dependent | rect. | 5 | 22 |
| 53 | 97.73 | humidity | 80 | 1440 | < th | dose independent | sin | 2 | NA |
| 69 | 98.07 | humidity | 70 | 720 | > th | dose dependent | rect. | 9 | 14 |
| 71 | 98.22 | humidity | 80 | 1440 | > th | dose independent | sin | 2 | NA |
| 120 | 99.17 | humidity | 80 | 720 | < th | dose independent | rect. | 8 | 13 |
| 126 | 99.23 | humidity | 80 | 10 | > th | dose independent | rect. | 6 | 14 |
| 135 | 99.35 | humidity | 80 | 720 | > th | dose independent | rect. | 8 | 14 |
| 160 | 99.75 | humidity | 70 | 10 | > th | dose dependent | sin | 21 | NA |
| 167 | 99.85 | humidity | 70 | 720 | > th | dose dependent | rect. | 4 | 19 |
| 195 | 100.09 | humidity | 90 | 1440 | < th | dose dependent | rect. | 9 | 7 |
| 209 | 100.24 | humidity | 80 | 720 | > th | dose independent | rect. | 4 | 18 |
| 228 | 100.38 | humidity | 70 | 720 | > th | dose dependent | sin | 21 | NA |
| 237 | 100.42 | humidity | 60 | 720 | > th | dose dependent | sin | 19 | NA |
| 250 | 100.51 | humidity | 70 | 90 | > th | dose dependent | rect. | 5 | 18 |
| 261 | 100.59 | humidity | 80 | 720 | > th | dose independent | sin | 22 | NA |
| 385 | 101.31 | humidity | 60 | 720 | > th | dose dependent | rect. | 9 | 22 |
| 445 | 101.62 | humidity | 50 | 1440 | > th | dose dependent | sin | 0 | NA |
| 473 | 101.71 | humidity | 90 | 1440 | < th | dose dependent | sin | 0 | NA |
| 545 | 101.93 | humidity | 50 | 1440 | > th | dose dependent | rect. | 13 | 1 |
| 786 | 102.67 | humidity | 90 | 1440 | < th | dose dependent | rect. | 13 | 1 |
| 929 | 103.09 | humidity | 70 | 720 | > th | dose independent | rect. | 13 | 18 |
| 946 | 103.14 | radiation | 1 | 720 | < th | dose independent | rect. | 7 | 18 |
| 953 | 103.16 | humidity | 70 | 720 | > th | dose independent | rect. | 13 | 10 |