Os11g0530600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Chalcone synthase C2 (EC 2.3.1.74) (Naringenin-chalcone synthase C2).


FiT-DB / Search/ Help/ Sample detail
|
Os11g0530600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Chalcone synthase C2 (EC 2.3.1.74) (Naringenin-chalcone synthase C2).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
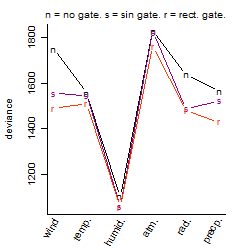
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1028.56 | 1.51 | 13.5 | -5.14 | 2.55 | -5.81 | 5.96 | -7.39 | -1.66 | 10.3 | 75 | 1302 | -- | -- |
| 1138.85 | 1.57 | 13.6 | -6.09 | 2.84 | -6.06 | 5.02 | -- | -1.9 | 10.3 | 75 | 1262 | -- | -- |
| 1156.17 | 1.58 | 13.5 | -4.86 | 2 | -5.28 | -- | -7.32 | -1.73 | 9.13 | 74.37 | 1450 | -- | -- |
| 1250.96 | 1.65 | 13.6 | -5.98 | 2.82 | -6.08 | -- | -- | -1.89 | 10.4 | 74.66 | 1256 | -- | -- |
| 1374.79 | 1.73 | 13.5 | -4.84 | -- | -5.31 | -- | -7.21 | -1.58 | -- | 74.45 | 1465 | -- | -- |
| 2262.47 | 2.23 | 13.4 | -5.24 | 2.06 | -- | 5.03 | -- | -1.14 | 8.93 | -- | -- | -- | -- |
| 2348.72 | 2.27 | 13.4 | -5.07 | 2.05 | -- | -- | -- | -1.14 | 8.84 | -- | -- | -- | -- |
| 1495.12 | 1.8 | 13.7 | -5.94 | -- | -5.66 | -- | -- | -1.88 | -- | 74.14 | 1448 | -- | -- |
| 1996.45 | 2.08 | 13.6 | -- | 3.07 | -6.82 | -- | -- | -1.37 | 10.2 | 82.69 | 2318 | -- | -- |
| 2588.66 | 2.38 | 13.5 | -5.07 | -- | -- | -- | -- | -1.13 | -- | -- | -- | -- | -- |
| 3026.46 | 2.57 | 13.3 | -- | 2.04 | -- | -- | -- | -0.576 | 9.02 | -- | -- | -- | -- |
| 2383.59 | 2.27 | 13.5 | -- | -- | -5.71 | -- | -- | -1.34 | -- | 75 | 2890 | -- | -- |
| 3266.66 | 2.67 | 13.4 | -- | -- | -- | -- | -- | -0.563 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 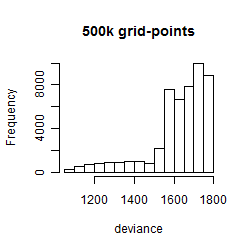
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1056.09 | humidity | 80 | 1440 | < th | dose independent | rect. | 6 | 14 |
| 5 | 1060.75 | humidity | 80 | 1440 | < th | dose independent | sin | 1 | NA |
| 7 | 1062.45 | humidity | 80 | 1440 | < th | dose independent | rect. | 7 | 8 |
| 9 | 1065.17 | humidity | 80 | 1440 | > th | dose independent | rect. | 6 | 14 |
| 12 | 1066.38 | humidity | 60 | 1440 | > th | dose dependent | rect. | 5 | 18 |
| 13 | 1066.60 | humidity | 70 | 1440 | > th | dose dependent | rect. | 6 | 15 |
| 15 | 1066.97 | humidity | 80 | 1440 | > th | dose independent | sin | 1 | NA |
| 65 | 1077.82 | humidity | 80 | 1440 | > th | dose independent | rect. | 6 | 9 |
| 66 | 1077.97 | humidity | 60 | 1440 | > th | dose dependent | sin | 1 | NA |
| 89 | 1080.39 | humidity | 70 | 1440 | > th | dose dependent | rect. | 6 | 9 |
| 194 | 1095.98 | humidity | 50 | 1440 | > th | dose dependent | rect. | 16 | 23 |
| 202 | 1096.69 | humidity | 90 | 1440 | < th | dose dependent | rect. | 16 | 22 |
| 219 | 1098.49 | humidity | 50 | 1440 | > th | dose dependent | rect. | 13 | 23 |
| 251 | 1101.90 | humidity | 50 | 1440 | > th | dose dependent | no | NA | NA |
| 407 | 1117.82 | humidity | 90 | 1440 | < th | dose dependent | rect. | 2 | 22 |
| 409 | 1117.86 | humidity | 90 | 1440 | < th | dose dependent | rect. | 24 | 23 |
| 422 | 1118.46 | humidity | 90 | 1440 | < th | dose dependent | rect. | 5 | 23 |
| 433 | 1119.08 | humidity | 90 | 1440 | < th | dose dependent | no | NA | NA |
| 599 | 1134.96 | humidity | 70 | 1440 | > th | dose independent | rect. | 18 | 23 |
| 746 | 1150.59 | humidity | 90 | 1440 | < th | dose dependent | sin | 2 | NA |
| 796 | 1156.69 | humidity | 70 | 1440 | > th | dose independent | rect. | 15 | 23 |
| 805 | 1157.07 | humidity | 70 | 1440 | > th | dose independent | rect. | 8 | 23 |
| 844 | 1158.29 | humidity | 70 | 1440 | > th | dose independent | rect. | 2 | 23 |
| 872 | 1158.38 | humidity | 70 | 1440 | > th | dose independent | no | NA | NA |