Os11g0497000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Ubiquitin activating enzyme-like protein (SUMO activating enzyme 1a).


FiT-DB / Search/ Help/ Sample detail
|
Os11g0497000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Ubiquitin activating enzyme-like protein (SUMO activating enzyme 1a).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
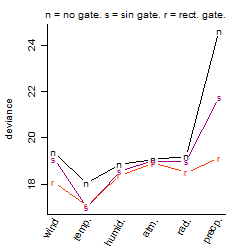
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17.64 | 0.197 | 11.4 | 0.0269 | 0.547 | 0.637 | 0.0857 | -0.356 | -0.0725 | 16.7 | 19.3 | 6542 | -- | -- |
| 17.95 | 0.197 | 11.4 | 0.0978 | 0.555 | 0.658 | -0.00858 | -- | -0.0677 | 16.6 | 19.2 | 6661 | -- | -- |
| 17.65 | 0.196 | 11.4 | 0.0268 | 0.548 | 0.638 | -- | -0.354 | -0.0724 | 16.7 | 19.31 | 6542 | -- | -- |
| 17.7 | 0.196 | 11.4 | 0.0486 | 0.545 | 0.766 | -- | -- | -0.062 | 16.7 | 19 | 6569 | -- | -- |
| 33.34 | 0.269 | 11.4 | 0.0183 | -- | 0.648 | -- | -0.343 | -0.0781 | -- | 19.08 | 6329 | -- | -- |
| 25.58 | 0.237 | 11.3 | 0.546 | 0.543 | -- | 0.0775 | -- | 0.127 | 16.8 | -- | -- | -- | -- |
| 25.6 | 0.237 | 11.3 | 0.546 | 0.544 | -- | -- | -- | 0.127 | 16.9 | -- | -- | -- | -- |
| 33.49 | 0.27 | 11.4 | 0.0431 | -- | 0.767 | -- | -- | -0.0867 | -- | 19.1 | 6372 | -- | -- |
| 17.71 | 0.196 | 11.4 | -- | 0.551 | 0.803 | -- | -- | -0.0736 | 16.7 | 19.01 | 6680 | -- | -- |
| 41.21 | 0.3 | 11.3 | 0.526 | -- | -- | -- | -- | 0.113 | -- | -- | -- | -- | -- |
| 33.47 | 0.271 | 11.4 | -- | 0.534 | -- | -- | -- | 0.0664 | 16.9 | -- | -- | -- | -- |
| 33.46 | 0.269 | 11.4 | -- | -- | 0.776 | -- | -- | -0.0824 | -- | 19.1 | 6350 | -- | -- |
| 48.51 | 0.325 | 11.4 | -- | -- | -- | -- | -- | 0.0549 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 17.00 | temperature | 15 | 270 | > th | dose dependent | sin | 7 | NA |
| 2 | 17.01 | temperature | 30 | 90 | < th | dose dependent | sin | 5 | NA |
| 4 | 17.07 | temperature | 30 | 270 | < th | dose dependent | rect. | 21 | 22 |
| 6 | 17.15 | temperature | 10 | 90 | > th | dose dependent | sin | 5 | NA |
| 8 | 17.16 | temperature | 30 | 270 | < th | dose dependent | sin | 7 | NA |
| 16 | 17.26 | temperature | 30 | 270 | < th | dose dependent | rect. | 23 | 22 |
| 28 | 17.43 | temperature | 20 | 4320 | < th | dose independent | rect. | 18 | 5 |
| 40 | 17.51 | temperature | 20 | 4320 | > th | dose independent | rect. | 18 | 5 |
| 50 | 17.54 | temperature | 25 | 4320 | < th | dose dependent | rect. | 19 | 1 |
| 110 | 17.67 | temperature | 25 | 720 | < th | dose dependent | rect. | 17 | 19 |
| 139 | 17.72 | temperature | 20 | 4320 | > th | dose independent | sin | 15 | NA |
| 147 | 17.73 | temperature | 20 | 4320 | < th | dose independent | sin | 15 | NA |
| 152 | 17.73 | temperature | 30 | 270 | < th | dose dependent | rect. | 23 | 16 |
| 222 | 17.80 | temperature | 25 | 4320 | < th | dose dependent | sin | 14 | NA |
| 251 | 17.82 | temperature | 10 | 4320 | > th | dose dependent | rect. | 19 | 6 |
| 268 | 17.83 | temperature | 10 | 4320 | > th | dose dependent | rect. | 19 | 2 |
| 414 | 17.90 | temperature | 30 | 270 | < th | dose dependent | rect. | 1 | 14 |
| 433 | 17.91 | temperature | 10 | 4320 | > th | dose dependent | rect. | 23 | 2 |
| 501 | 17.93 | temperature | 15 | 270 | > th | dose dependent | rect. | 17 | 22 |
| 551 | 17.95 | temperature | 25 | 270 | < th | dose dependent | rect. | 23 | 12 |
| 636 | 17.98 | temperature | 10 | 4320 | > th | dose dependent | sin | 13 | NA |
| 770 | 18.01 | temperature | 30 | 10 | < th | dose dependent | rect. | 3 | 23 |
| 824 | 18.03 | temperature | 25 | 4320 | < th | dose dependent | no | NA | NA |
| 892 | 18.04 | temperature | 25 | 4320 | < th | dose dependent | rect. | 2 | 23 |
| 930 | 18.05 | temperature | 25 | 4320 | < th | dose dependent | rect. | 23 | 23 |
| 969 | 18.06 | temperature | 30 | 90 | < th | dose dependent | rect. | 15 | 23 |
| 973 | 18.07 | wind | 1 | 43200 | > th | dose independent | rect. | 24 | 2 |