Os11g0487100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.

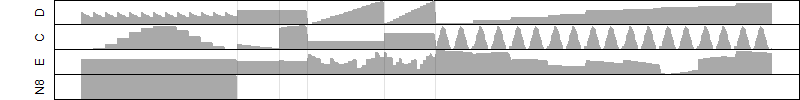
FiT-DB / Search/ Help/ Sample detail
|
Os11g0487100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
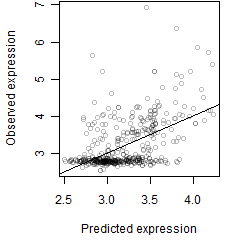
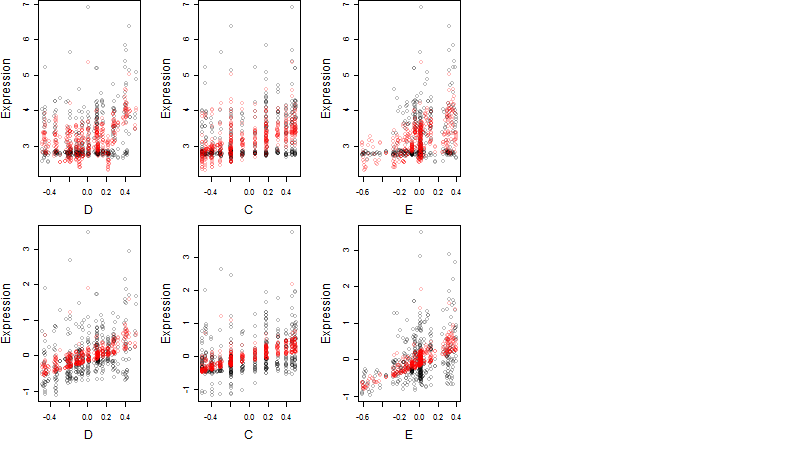
Dependence on each variable
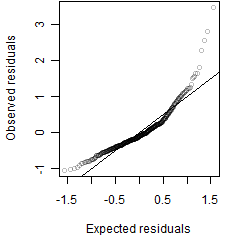
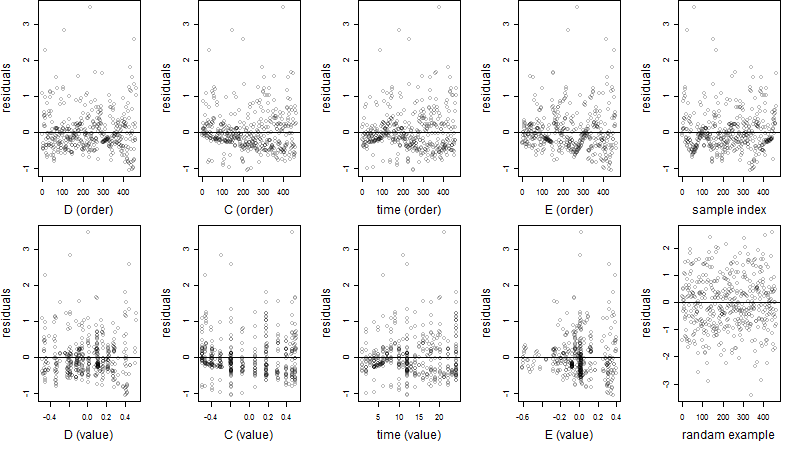
Residual plot
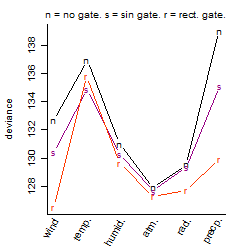
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 127.96 | 0.531 | 3.23 | 0.733 | 0.607 | 0.959 | 1.84 | 0.789 | -0.0895 | 19.3 | 997.9 | 10861 | -- | -- |
| 128.66 | 0.528 | 3.22 | 0.88 | 0.601 | 1.02 | 1.81 | -- | -0.0714 | 19.2 | 997 | 10900 | -- | -- |
| 136.91 | 0.545 | 3.23 | 0.692 | 0.612 | 0.955 | -- | 0.825 | -0.0837 | 19.5 | 997.7 | 10794 | -- | -- |
| 137.72 | 0.547 | 3.22 | 0.845 | 0.611 | 1.03 | -- | -- | -0.0621 | 19.5 | 996.2 | 10880 | -- | -- |
| 156.64 | 0.583 | 3.22 | 0.665 | -- | 0.97 | -- | 0.941 | -0.102 | -- | 997.8 | 10175 | -- | -- |
| 150.2 | 0.575 | 3.22 | 0.651 | 0.615 | -- | 1.85 | -- | -0.0692 | 19.2 | -- | -- | -- | -- |
| 159.24 | 0.591 | 3.22 | 0.611 | 0.617 | -- | -- | -- | -0.0662 | 19.4 | -- | -- | -- | -- |
| 157.51 | 0.585 | 3.2 | 0.838 | -- | 1.05 | -- | -- | -0.0704 | -- | 996.4 | 11039 | -- | -- |
| 155.01 | 0.58 | 3.23 | -- | 0.611 | 0.843 | -- | -- | -0.137 | 19.3 | 999.4 | 17298 | -- | -- |
| 179.56 | 0.626 | 3.2 | 0.601 | -- | -- | -- | -- | -0.0761 | -- | -- | -- | -- | -- |
| 169.07 | 0.608 | 3.23 | -- | 0.612 | -- | -- | -- | -0.134 | 19.5 | -- | -- | -- | -- |
| 174.79 | 0.616 | 3.22 | -- | -- | 0.863 | -- | -- | -0.141 | -- | 999.9 | 17191 | -- | -- |
| 189.09 | 0.642 | 3.22 | -- | -- | -- | -- | -- | -0.143 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance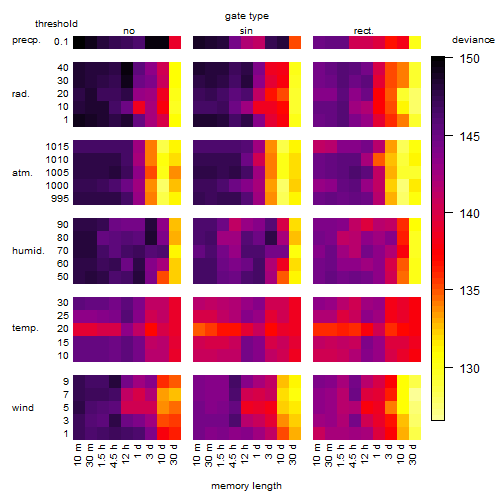
|
Histogram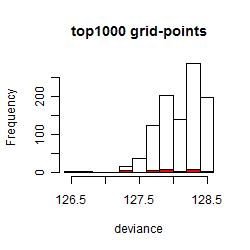 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 126.53 | wind | 7 | 43200 | < th | dose independent | rect. | 14 | 1 |
| 2 | 126.69 | wind | 7 | 43200 | > th | dose independent | rect. | 14 | 1 |
| 3 | 127.30 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 15 | 1 |
| 4 | 127.30 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 13 | 3 |
| 6 | 127.33 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 13 | 1 |
| 11 | 127.37 | wind | 5 | 43200 | > th | dose dependent | rect. | 14 | 1 |
| 56 | 127.63 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 19 | 1 |
| 68 | 127.66 | atmosphere | 1000 | 14400 | > th | dose dependent | sin | 20 | NA |
| 108 | 127.74 | radiation | 20 | 43200 | > th | dose dependent | rect. | 6 | 2 |
| 152 | 127.78 | radiation | 20 | 43200 | < th | dose independent | rect. | 7 | 1 |
| 290 | 127.90 | atmosphere | 1000 | 14400 | > th | dose dependent | no | NA | NA |
| 305 | 127.91 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 19 | 23 |
| 318 | 127.93 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 17 | 23 |
| 351 | 127.99 | radiation | 20 | 43200 | > th | dose independent | rect. | 17 | 14 |
| 352 | 127.99 | radiation | 10 | 43200 | < th | dose dependent | rect. | 13 | 1 |
| 361 | 128.00 | radiation | 20 | 43200 | > th | dose independent | rect. | 6 | 1 |
| 375 | 128.00 | wind | 7 | 43200 | > th | dose dependent | rect. | 16 | 3 |
| 395 | 128.04 | atmosphere | 1015 | 14400 | < th | dose dependent | rect. | 19 | 1 |
| 436 | 128.11 | atmosphere | 1015 | 14400 | < th | dose dependent | rect. | 17 | 1 |
| 462 | 128.14 | atmosphere | 1015 | 14400 | < th | dose dependent | rect. | 15 | 5 |
| 518 | 128.21 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 4 | 1 |
| 575 | 128.25 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 23 | 6 |
| 590 | 128.27 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 23 | 1 |
| 614 | 128.28 | radiation | 20 | 43200 | < th | dose independent | rect. | 18 | 14 |
| 633 | 128.29 | radiation | 20 | 43200 | < th | dose independent | rect. | 21 | 11 |
| 792 | 128.40 | atmosphere | 1015 | 14400 | < th | dose dependent | sin | 18 | NA |
| 911 | 128.47 | atmosphere | 1000 | 14400 | > th | dose dependent | rect. | 7 | 1 |