Os11g0272900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Zinc finger, SWIM-type domain containing protein.

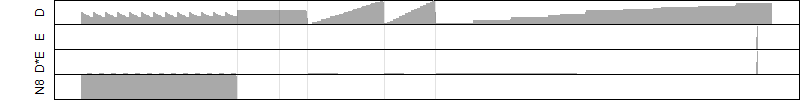
FiT-DB / Search/ Help/ Sample detail
|
Os11g0272900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Zinc finger, SWIM-type domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

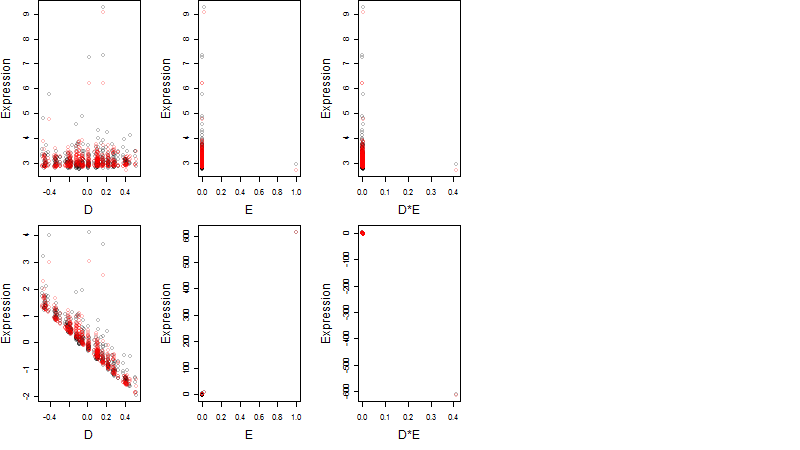
Dependence on each variable
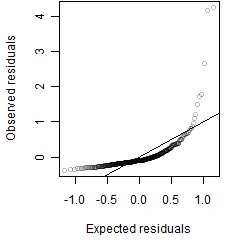
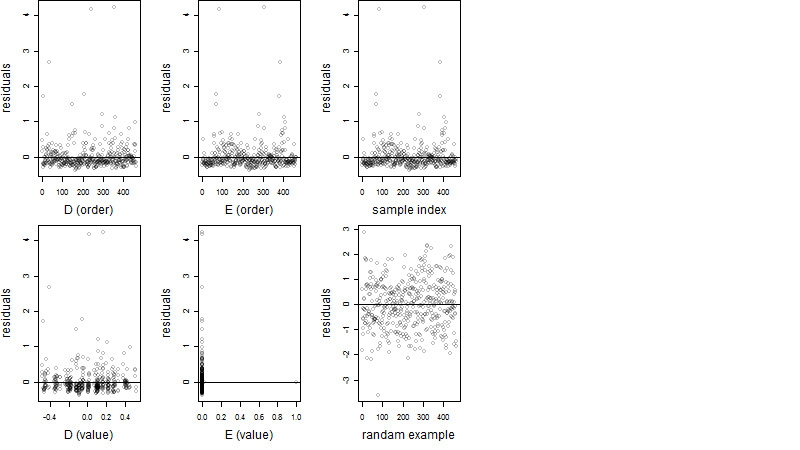
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose independent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 75.14 | 0.404 | 4.47 | -3.26 | 0.0941 | 615 | -0.355 | -1500 | -0.0239 | 2.34 | 50 | 90 | 13 | 1 |
| 112.81 | 0.498 | 3.13 | 0.0642 | 0.0203 | -0.292 | -0.497 | -- | -0.0363 | 0.866 | 50 | 90 | 13 | 1 |
| 75.46 | 0.405 | 4.46 | -3.25 | 0.111 | 615 | -- | -1500 | -0.0225 | 4.17 | 49.95 | 91 | 13 | 1 |
| 113.58 | 0.5 | 3.12 | 0.0776 | 0.0646 | -0.318 | -- | -- | -0.0353 | 5.4 | 51 | 58 | 14.2 | 2.18 |
| 76.12 | 0.406 | 4.46 | -3.25 | -- | 615 | -- | -1500 | -0.0194 | -- | 49.84 | 91 | 13 | 0.997 |
| 112.9 | 0.498 | 3.13 | 0.0603 | 0.0223 | -- | -0.484 | -- | -0.0359 | 0.849 | -- | -- | -- | -- |
| 113.68 | 0.499 | 3.12 | 0.0767 | 0.0675 | -- | -- | -- | -0.0346 | 5.36 | -- | -- | -- | -- |
| 112.32 | 0.494 | 3.13 | 0.0672 | -- | -0.142 | -- | -- | 0.024 | -- | 106.1 | 41 | 14.3 | 5.52 |
| 95.5 | 0.458 | 3.12 | -- | 0.112 | 3.05 | -- | -- | -0.0271 | 4.16 | 53 | 12 | 12.9 | 2.88 |
| 113.92 | 0.499 | 3.12 | 0.079 | -- | -- | -- | -- | -0.0329 | -- | -- | -- | -- | -- |
| 113.84 | 0.499 | 3.13 | -- | 0.0688 | -- | -- | -- | -0.0431 | 5.32 | -- | -- | -- | -- |
| 96.18 | 0.458 | 3.12 | -- | -- | 3 | -- | -- | -0.0249 | -- | 53 | 12 | 12.9 | 2.88 |
| 114.09 | 0.499 | 3.13 | -- | -- | -- | -- | -- | -0.0417 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 75.14 | humidity | 50 | 90 | < th | dose independent | rect. | 13 | 1 |
| 3 | 75.14 | humidity | 50 | 90 | < th | dose dependent | rect. | 13 | 1 |
| 5 | 75.20 | atmosphere | 1000 | 90 | < th | dose independent | rect. | 12 | 1 |
| 6 | 75.20 | atmosphere | 1000 | 90 | < th | dose dependent | rect. | 12 | 2 |
| 7 | 75.21 | atmosphere | 1000 | 30 | < th | dose dependent | rect. | 8 | 7 |
| 69 | 85.70 | temperature | 30 | 30 | > th | dose dependent | rect. | 13 | 1 |
| 77 | 86.02 | atmosphere | 1000 | 270 | < th | dose independent | rect. | 9 | 5 |
| 126 | 88.90 | atmosphere | 1000 | 270 | < th | dose dependent | rect. | 9 | 2 |
| 141 | 89.68 | atmosphere | 1000 | 10 | < th | dose dependent | sin | 1 | NA |
| 164 | 90.75 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 2 | 13 |
| 174 | 91.18 | atmosphere | 1000 | 720 | < th | dose dependent | rect. | 7 | 2 |
| 311 | 94.23 | atmosphere | 1000 | 720 | < th | dose independent | rect. | 3 | 7 |
| 392 | 94.73 | atmosphere | 1000 | 720 | < th | dose independent | rect. | 5 | 4 |
| 412 | 94.87 | atmosphere | 1000 | 720 | < th | dose independent | rect. | 2 | 11 |
| 455 | 95.48 | atmosphere | 1000 | 720 | < th | dose dependent | sin | 6 | NA |
| 495 | 95.66 | atmosphere | 1000 | 720 | > th | dose independent | rect. | 3 | 8 |
| 519 | 95.89 | temperature | 30 | 270 | > th | dose dependent | rect. | 9 | 5 |
| 678 | 97.19 | temperature | 30 | 270 | > th | dose dependent | sin | 8 | NA |
| 738 | 97.56 | atmosphere | 1000 | 270 | < th | dose independent | sin | 2 | NA |
| 744 | 97.65 | temperature | 30 | 30 | > th | dose independent | rect. | 13 | 1 |
| 753 | 97.74 | atmosphere | 1000 | 270 | > th | dose independent | sin | 2 | NA |
| 819 | 98.21 | temperature | 30 | 270 | > th | dose dependent | rect. | 10 | 1 |
| 1000 | 99.54 | atmosphere | 1000 | 720 | < th | dose independent | sin | 7 | NA |