Os11g0175000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF1618 domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os11g0175000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF1618 domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
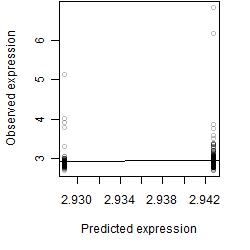 __
__
Dependence on each variable

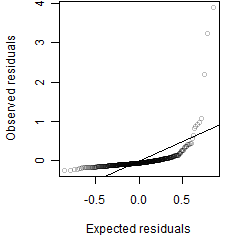
Residual plot

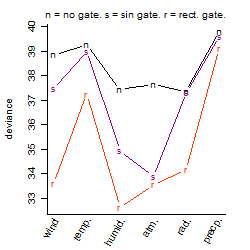
Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 35.78 | 0.281 | 2.93 | 0.0568 | 0.106 | 0.396 | 0.262 | -1.72 | 0.013 | 9.07 | 50 | 15 | -- | -- |
| 36.99 | 0.283 | 2.94 | 0.0632 | 0.112 | 0.811 | 0.0676 | -- | 0.0112 | 8.15 | 54.26 | 5 | -- | -- |
| 35.74 | 0.281 | 2.93 | 0.042 | 0.113 | 0.329 | -- | -2.34 | 0.0103 | 8.14 | 50 | 14 | -- | -- |
| 37 | 0.283 | 2.93 | 0.0538 | 0.111 | 0.849 | -- | -- | 0.00862 | 8.08 | 53.89 | 4 | -- | -- |
| 36.43 | 0.283 | 2.93 | 0.0455 | -- | 0.376 | -- | -2.34 | 0.0131 | -- | 50 | 14 | -- | -- |
| 39.92 | 0.296 | 2.94 | 0.00519 | 0.141 | -- | -0.00161 | -- | -0.0142 | 9.21 | -- | -- | -- | -- |
| 39.92 | 0.296 | 2.94 | 0.00513 | 0.141 | -- | -- | -- | -0.0142 | 9.21 | -- | -- | -- | -- |
| 37.66 | 0.286 | 2.94 | 0.0572 | -- | 0.892 | -- | -- | 0.0112 | -- | 53.76 | 4 | -- | -- |
| 37.07 | 0.284 | 2.94 | -- | 0.112 | 0.827 | -- | -- | 0.00231 | 8.07 | 53.76 | 4 | -- | -- |
| 41.08 | 0.299 | 2.94 | 0.00461 | -- | -- | -- | -- | -0.0135 | -- | -- | -- | -- | -- |
| 39.92 | 0.296 | 2.94 | -- | 0.141 | -- | -- | -- | -0.0147 | 9.21 | -- | -- | -- | -- |
| 37.75 | 0.286 | 2.94 | -- | -- | 0.872 | -- | -- | 0.00456 | -- | 53.76 | 4 | -- | -- |
| 41.08 | 0.299 | 2.94 | -- | -- | -- | -- | -- | -0.014 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram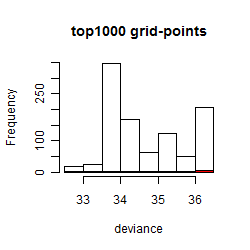 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 32.64 | humidity | 70 | 4320 | > th | dose independent | rect. | 20 | 10 |
| 2 | 32.70 | humidity | 70 | 4320 | > th | dose independent | rect. | 21 | 1 |
| 6 | 32.72 | humidity | 70 | 4320 | > th | dose independent | rect. | 20 | 3 |
| 20 | 33.16 | humidity | 50 | 4320 | > th | dose independent | rect. | 16 | 1 |
| 21 | 33.23 | humidity | 70 | 4320 | < th | dose independent | rect. | 20 | 1 |
| 31 | 33.23 | humidity | 70 | 4320 | < th | dose dependent | rect. | 20 | 1 |
| 43 | 33.55 | atmosphere | 1010 | 10 | > th | dose dependent | rect. | 12 | 1 |
| 75 | 33.59 | wind | 9 | 4320 | > th | dose dependent | rect. | 14 | 1 |
| 154 | 33.89 | atmosphere | 1010 | 30 | > th | dose dependent | sin | 23 | NA |
| 426 | 34.16 | radiation | 1 | 270 | < th | dose independent | rect. | 8 | 1 |
| 428 | 34.16 | radiation | 1 | 270 | < th | dose dependent | rect. | 8 | 1 |
| 568 | 34.62 | wind | 9 | 4320 | > th | dose independent | rect. | 14 | 1 |
| 615 | 34.95 | humidity | 50 | 10 | > th | dose independent | sin | 5 | NA |
| 722 | 35.35 | wind | 9 | 4320 | < th | dose independent | rect. | 14 | 1 |
| 761 | 35.69 | humidity | 60 | 270 | < th | dose independent | rect. | 10 | 1 |
| 796 | 36.01 | atmosphere | 1010 | 10 | > th | dose independent | rect. | 11 | 1 |
| 864 | 36.02 | humidity | 70 | 4320 | < th | dose dependent | rect. | 19 | 14 |
| 922 | 36.19 | humidity | 70 | 4320 | > th | dose independent | rect. | 6 | 1 |
| 933 | 36.20 | humidity | 60 | 14400 | < th | dose independent | rect. | 6 | 1 |
| 938 | 36.20 | radiation | 20 | 14400 | > th | dose independent | rect. | 17 | 1 |
| 996 | 36.20 | humidity | 60 | 14400 | < th | dose dependent | rect. | 6 | 1 |
| 1000 | 36.20 | radiation | 20 | 14400 | > th | dose dependent | rect. | 17 | 1 |