Os11g0170000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Amidase family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os11g0170000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Amidase family protein.
|
|
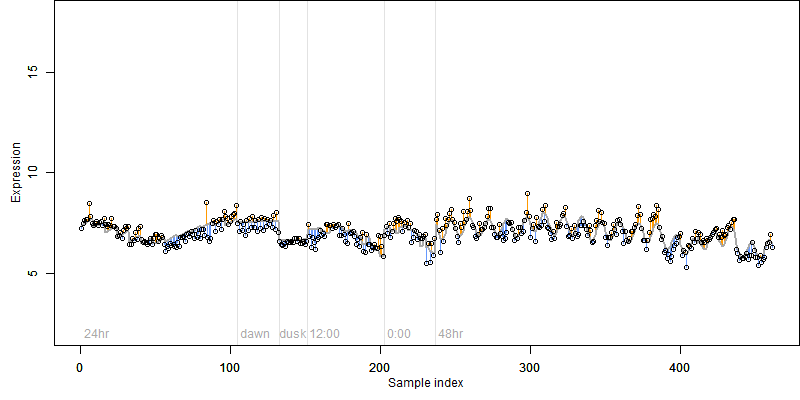
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

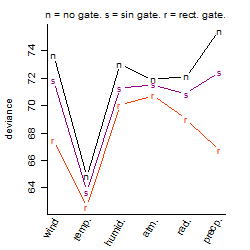
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 61.98 | 0.37 | 7.04 | -1.23 | 0.978 | -0.752 | -0.179 | -0.623 | -0.259 | 5.92 | 22.49 | 1885 | -- | -- |
| 63.47 | 0.371 | 7.05 | -1.33 | 0.952 | -0.749 | -0.282 | -- | -0.327 | 5.97 | 22.69 | 1857 | -- | -- |
| 62.05 | 0.367 | 7.04 | -1.23 | 0.978 | -0.753 | -- | -0.633 | -0.258 | 5.94 | 22.48 | 1897 | -- | -- |
| 62.97 | 0.37 | 7.06 | -1.44 | 0.977 | -0.749 | -- | -- | -0.313 | 5.98 | 22.6 | 1917 | -- | -- |
| 107.91 | 0.484 | 7.06 | -1.19 | -- | -0.777 | -- | -0.629 | -0.243 | -- | 22.49 | 1403 | -- | -- |
| 85.29 | 0.433 | 7 | -1.16 | 0.966 | -- | -0.242 | -- | -0.0419 | 5.82 | -- | -- | -- | -- |
| 85.44 | 0.433 | 7 | -1.16 | 0.965 | -- | -- | -- | -0.0409 | 5.84 | -- | -- | -- | -- |
| 108.01 | 0.484 | 7.09 | -1.49 | -- | -0.855 | -- | -- | -0.323 | -- | 24.04 | 2776 | -- | -- |
| 112.4 | 0.494 | 7.02 | -- | 0.939 | -0.456 | -- | -- | -0.089 | 6.12 | 22.8 | 1193 | -- | -- |
| 133.73 | 0.54 | 7.02 | -1.13 | -- | -- | -- | -- | -0.0184 | -- | -- | -- | -- | -- |
| 120.78 | 0.514 | 6.97 | -- | 0.947 | -- | -- | -- | 0.0881 | 5.9 | -- | -- | -- | -- |
| 156.97 | 0.584 | 7.02 | -- | -- | -0.475 | -- | -- | -0.0483 | -- | 22.67 | 1314 | -- | -- |
| 167.33 | 0.604 | 6.99 | -- | -- | -- | -- | -- | 0.107 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram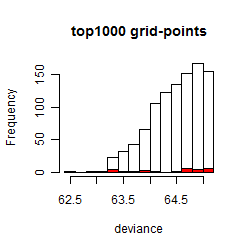 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 62.58 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 19 |
| 4 | 63.23 | temperature | 15 | 720 | > th | dose dependent | rect. | 18 | 14 |
| 6 | 63.26 | temperature | 20 | 1440 | > th | dose dependent | rect. | 23 | 6 |
| 9 | 63.32 | temperature | 20 | 1440 | > th | dose dependent | rect. | 20 | 9 |
| 10 | 63.33 | temperature | 20 | 1440 | > th | dose dependent | rect. | 1 | 2 |
| 31 | 63.43 | temperature | 30 | 720 | < th | dose dependent | rect. | 15 | 14 |
| 81 | 63.69 | temperature | 10 | 720 | > th | dose dependent | sin | 12 | NA |
| 120 | 63.88 | temperature | 30 | 1440 | < th | dose dependent | sin | 14 | NA |
| 124 | 63.89 | temperature | 20 | 1440 | > th | dose dependent | sin | 13 | NA |
| 201 | 64.07 | temperature | 25 | 720 | < th | dose dependent | rect. | 9 | 20 |
| 478 | 64.54 | temperature | 20 | 1440 | > th | dose dependent | rect. | 20 | 1 |
| 572 | 64.67 | temperature | 30 | 1440 | < th | dose dependent | rect. | 16 | 3 |
| 587 | 64.69 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 1 |
| 592 | 64.70 | temperature | 30 | 1440 | < th | dose dependent | rect. | 23 | 1 |
| 646 | 64.77 | temperature | 25 | 720 | < th | dose dependent | rect. | 5 | 22 |
| 671 | 64.79 | temperature | 25 | 720 | < th | dose dependent | rect. | 1 | 23 |
| 681 | 64.81 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 695 | 64.82 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 745 | 64.88 | temperature | 25 | 720 | < th | dose dependent | rect. | 17 | 23 |
| 804 | 64.96 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 1 |
| 862 | 65.02 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 20 |
| 875 | 65.06 | temperature | 25 | 1440 | < th | dose independent | rect. | 12 | 15 |
| 880 | 65.06 | temperature | 25 | 720 | < th | dose dependent | rect. | 23 | 23 |
| 922 | 65.10 | temperature | 25 | 720 | < th | dose dependent | rect. | 21 | 23 |
| 963 | 65.14 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 5 |
| 996 | 65.19 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 2 |