Os10g0406100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to PRLI-interacting factor F (Fragment).


FiT-DB / Search/ Help/ Sample detail
|
Os10g0406100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to PRLI-interacting factor F (Fragment).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
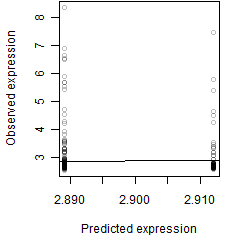
Dependence on each variable

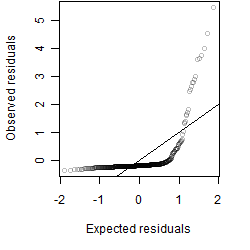
Residual plot

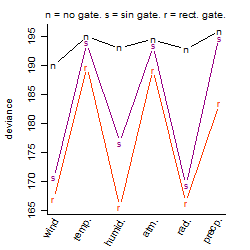
Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = > th, dose dependency = dose dependent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 166.14 | 0.606 | 2.88 | 0.0716 | 0.151 | 1.95 | -0.066 | -9.32 | 0.0553 | 2.85 | 45.07 | 15 | 5.69 | -- |
| 178.84 | 0.623 | 2.88 | 0.0834 | 0.168 | 3.35 | 0.167 | -- | 0.0655 | 2.42 | 47.17 | 25 | 5.67 | -- |
| 165.21 | 0.603 | 2.89 | 0.0143 | 0.158 | 2.49 | -- | -13.7 | 0.0466 | 2.41 | 45.49 | 15 | 5.67 | -- |
| 168.5 | 0.605 | 2.9 | 0.0585 | 0.236 | 5.13 | -- | -- | 0.00429 | 4.35 | 59.79 | 23 | 6.21 | -- |
| 166.65 | 0.605 | 2.88 | 0.0208 | -- | 2.39 | -- | -13.7 | 0.0497 | -- | 45.49 | 15 | 5.67 | -- |
| 196.68 | 0.657 | 2.89 | 0.0475 | 0.0886 | -- | -0.601 | -- | 0.0243 | 7.38 | -- | -- | -- | -- |
| 197.17 | 0.658 | 2.89 | 0.0304 | 0.13 | -- | -- | -- | 0.0234 | 4.47 | -- | -- | -- | -- |
| 168.52 | 0.605 | 2.88 | 0.159 | -- | 5.41 | -- | -- | 0.0644 | -- | 59.73 | 23 | 6.21 | -- |
| 168.03 | 0.604 | 2.89 | -- | 0.181 | 5.43 | -- | -- | 0.036 | 2.83 | 59.66 | 22 | 6.25 | -- |
| 198.07 | 0.658 | 2.89 | 0.0354 | -- | -- | -- | -- | 0.0269 | -- | -- | -- | -- | -- |
| 197.2 | 0.657 | 2.89 | -- | 0.13 | -- | -- | -- | 0.0201 | 4.47 | -- | -- | -- | -- |
| 168.65 | 0.605 | 2.88 | -- | -- | 5.49 | -- | -- | 0.0666 | -- | 59.64 | 20 | 6.22 | -- |
| 198.11 | 0.657 | 2.89 | -- | -- | -- | -- | -- | 0.0229 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram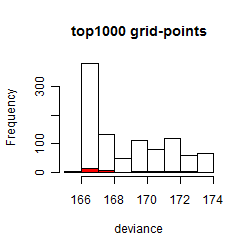 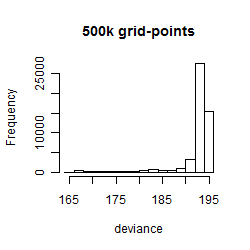
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 165.53 | humidity | 60 | 1440 | > th | dose independent | rect. | 6 | 3 |
| 3 | 166.12 | humidity | 60 | 1440 | > th | dose independent | rect. | 23 | 10 |
| 4 | 166.22 | radiation | 40 | 30 | > th | dose dependent | rect. | 9 | 1 |
| 25 | 166.35 | humidity | 70 | 30 | < th | dose dependent | rect. | 9 | 1 |
| 67 | 166.67 | humidity | 60 | 90 | < th | dose dependent | rect. | 18 | 16 |
| 70 | 166.70 | humidity | 60 | 90 | < th | dose independent | rect. | 18 | 15 |
| 74 | 166.76 | humidity | 60 | 30 | < th | dose independent | rect. | 17 | 17 |
| 82 | 166.78 | humidity | 60 | 1440 | < th | dose dependent | rect. | 8 | 2 |
| 96 | 166.82 | humidity | 60 | 1440 | < th | dose independent | rect. | 8 | 1 |
| 132 | 166.86 | wind | 1 | 10 | < th | dose dependent | rect. | 8 | 4 |
| 142 | 166.90 | humidity | 60 | 30 | < th | dose independent | rect. | 9 | 1 |
| 287 | 166.93 | humidity | 60 | 270 | < th | dose independent | rect. | 8 | 1 |
| 316 | 166.93 | humidity | 60 | 270 | < th | dose dependent | rect. | 8 | 1 |
| 380 | 167.00 | wind | 1 | 10 | < th | dose dependent | rect. | 8 | 10 |
| 384 | 167.01 | wind | 1 | 10 | < th | dose dependent | rect. | 8 | 8 |
| 401 | 167.22 | wind | 1 | 90 | < th | dose dependent | rect. | 9 | 2 |
| 402 | 167.25 | wind | 1 | 10 | < th | dose independent | rect. | 8 | 2 |
| 461 | 167.39 | humidity | 60 | 1440 | > th | dose independent | rect. | 20 | 13 |
| 488 | 167.58 | wind | 1 | 10 | < th | dose independent | rect. | 8 | 10 |
| 493 | 167.59 | wind | 1 | 10 | < th | dose independent | rect. | 8 | 8 |
| 569 | 169.28 | radiation | 40 | 10 | > th | dose dependent | sin | 12 | NA |
| 575 | 169.52 | radiation | 40 | 30 | > th | dose independent | rect. | 9 | 1 |
| 709 | 170.63 | wind | 1 | 10 | < th | dose independent | sin | 21 | NA |
| 840 | 171.92 | wind | 1 | 10 | < th | dose dependent | sin | 21 | NA |
| 879 | 172.22 | wind | 1 | 10 | > th | dose independent | sin | 22 | NA |
| 893 | 172.33 | radiation | 40 | 30 | > th | dose independent | sin | 11 | NA |