Os09g0506700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Cyclin-like F-box domain containing protein.

FiT-DB / Search/ Help/ Sample detail
|
Os09g0506700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Cyclin-like F-box domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

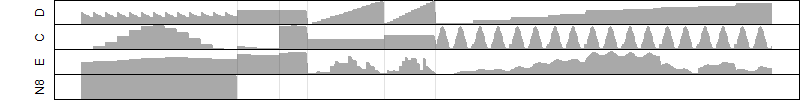
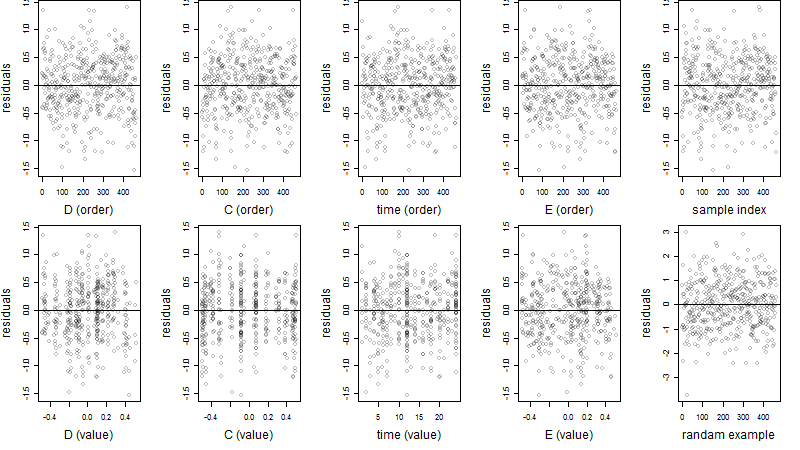
Dependence on each variable

Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 111.66 | 0.496 | 4.96 | 1.91 | 0.726 | 2.05 | -0.657 | 1.77 | 0.227 | 19 | 20.28 | 2102 | -- | -- |
| 115.09 | 0.5 | 4.98 | 1.55 | 0.627 | 2.03 | -0.561 | -- | 0.182 | 18.8 | 21.01 | 2132 | -- | -- |
| 112.59 | 0.494 | 4.96 | 1.9 | 0.751 | 2.09 | -- | 1.72 | 0.221 | 18.7 | 19.94 | 2185 | -- | -- |
| 114.36 | 0.498 | 5.01 | 1.45 | 0.737 | 2.12 | -- | -- | 0.101 | 18.6 | 20.9 | 2190 | -- | -- |
| 139.6 | 0.55 | 4.94 | 1.9 | -- | 2.19 | -- | 1.83 | 0.204 | -- | 20.6 | 2071 | -- | -- |
| 239.23 | 0.725 | 4.92 | 2.19 | 0.964 | -- | -0.365 | -- | 0.564 | 19.6 | -- | -- | -- | -- |
| 239.61 | 0.725 | 4.92 | 2.2 | 0.964 | -- | -- | -- | 0.563 | 19.6 | -- | -- | -- | -- |
| 139.96 | 0.551 | 5 | 1.42 | -- | 2.22 | -- | -- | 0.0703 | -- | 21.52 | 2068 | -- | -- |
| 143.02 | 0.557 | 5.13 | -- | 0.849 | 2.53 | -- | -- | -0.301 | 19.2 | 17.07 | 8016 | -- | -- |
| 289.55 | 0.795 | 4.9 | 2.19 | -- | -- | -- | -- | 0.549 | -- | -- | -- | -- | -- |
| 367.18 | 0.896 | 4.97 | -- | 0.947 | -- | -- | -- | 0.318 | 19.8 | -- | -- | -- | -- |
| 177.12 | 0.62 | 5.11 | -- | -- | 2.38 | -- | -- | -0.416 | -- | 17.69 | 25231 | -- | -- |
| 415.7 | 0.952 | 4.95 | -- | -- | -- | -- | -- | 0.306 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance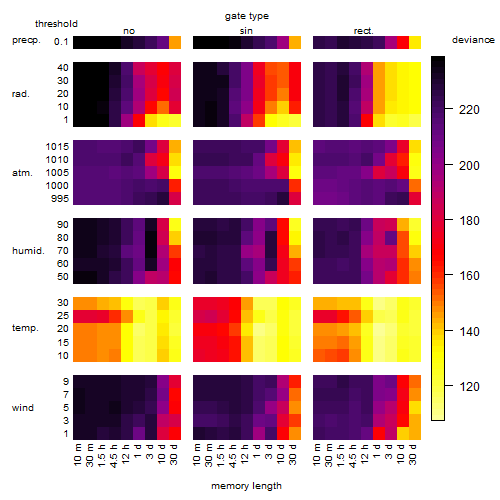
|
Histogram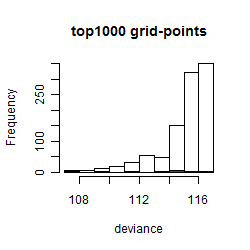 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 107.24 | temperature | 20 | 1440 | > th | dose dependent | rect. | 5 | 4 |
| 4 | 107.95 | temperature | 20 | 1440 | > th | dose dependent | rect. | 3 | 7 |
| 13 | 109.52 | temperature | 30 | 1440 | < th | dose dependent | rect. | 5 | 5 |
| 18 | 109.85 | temperature | 30 | 1440 | < th | dose dependent | rect. | 6 | 2 |
| 45 | 111.12 | temperature | 20 | 1440 | > th | dose dependent | sin | 6 | NA |
| 60 | 111.66 | temperature | 20 | 1440 | > th | dose dependent | rect. | 18 | 18 |
| 138 | 113.24 | temperature | 20 | 4320 | > th | dose dependent | rect. | 22 | 1 |
| 149 | 113.49 | temperature | 10 | 4320 | > th | dose dependent | rect. | 22 | 1 |
| 156 | 113.57 | temperature | 30 | 4320 | < th | dose dependent | rect. | 21 | 2 |
| 266 | 114.80 | temperature | 20 | 1440 | > th | dose dependent | no | NA | NA |
| 277 | 114.85 | temperature | 20 | 1440 | > th | dose dependent | rect. | 1 | 23 |
| 293 | 114.90 | temperature | 20 | 1440 | > th | dose dependent | rect. | 22 | 23 |
| 294 | 114.92 | temperature | 30 | 4320 | < th | dose dependent | rect. | 19 | 16 |
| 394 | 115.25 | temperature | 30 | 1440 | < th | dose dependent | rect. | 16 | 20 |
| 517 | 115.62 | temperature | 30 | 1440 | < th | dose dependent | sin | 5 | NA |
| 836 | 116.40 | temperature | 30 | 4320 | < th | dose dependent | sin | 9 | NA |
| 887 | 116.48 | temperature | 30 | 1440 | < th | dose dependent | rect. | 3 | 21 |