Os09g0495200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.
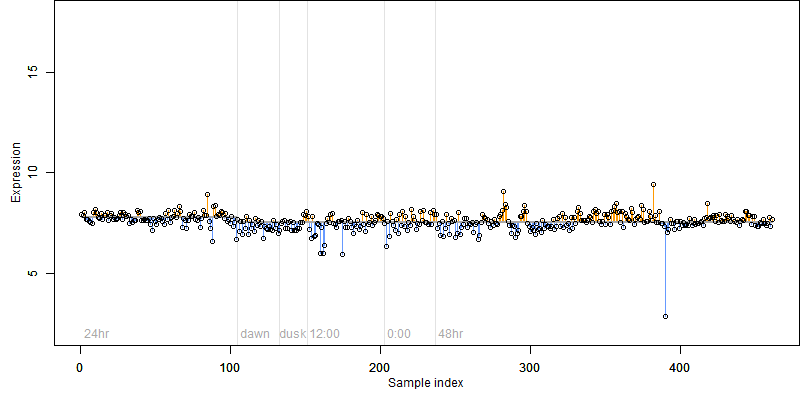

FiT-DB / Search/ Help/ Sample detail
|
Os09g0495200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
Dependence on each variable

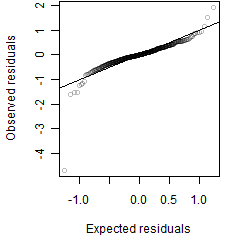
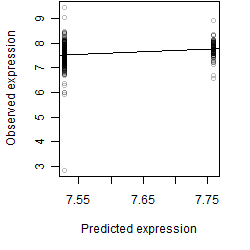
Residual plot

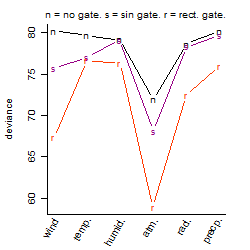
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 71.64 | 0.398 | 7.92 | -1.96 | 0.226 | 14 | -0.296 | -82 | 0.256 | 2.45 | 999.7 | 275 | -- | -- |
| 80.84 | 0.422 | 7.52 | 0.306 | 0.246 | 0.197 | -0.215 | -- | 0.267 | 2.36 | 999.7 | 275 | -- | -- |
| 71.64 | 0.394 | 7.9 | -1.8 | 0.237 | 14.3 | -- | -84.2 | 0.248 | 2.06 | 999.3 | 265 | -- | -- |
| 80.93 | 0.422 | 7.52 | 0.311 | 0.244 | 0.23 | -- | -- | 0.268 | 2.19 | 999.3 | 265 | -- | -- |
| 73.97 | 0.401 | 8.03 | -2.62 | -- | 22.3 | -- | -131 | 0.261 | -- | 998.9 | 227 | -- | -- |
| 81.15 | 0.422 | 7.52 | 0.321 | 0.245 | -- | -0.218 | -- | 0.261 | 2.35 | -- | -- | -- | -- |
| 81.32 | 0.422 | 7.52 | 0.327 | 0.243 | -- | -- | -- | 0.262 | 2.18 | -- | -- | -- | -- |
| 84.37 | 0.43 | 7.52 | 0.321 | -- | 0.269 | -- | -- | 0.275 | -- | 998.9 | 227 | -- | -- |
| 82.6 | 0.423 | 7.52 | -- | 0.259 | 0.52 | -- | -- | 0.245 | 2.31 | 996.2 | 746 | -- | -- |
| 84.85 | 0.43 | 7.52 | 0.337 | -- | -- | -- | -- | 0.268 | -- | -- | -- | -- | -- |
| 84.13 | 0.429 | 7.53 | -- | 0.25 | -- | -- | -- | 0.226 | 2.21 | -- | -- | -- | -- |
| 85.91 | 0.432 | 7.53 | -- | -- | 0.592 | -- | -- | 0.243 | -- | 996.4 | 758 | -- | -- |
| 87.85 | 0.437 | 7.53 | -- | -- | -- | -- | -- | 0.231 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance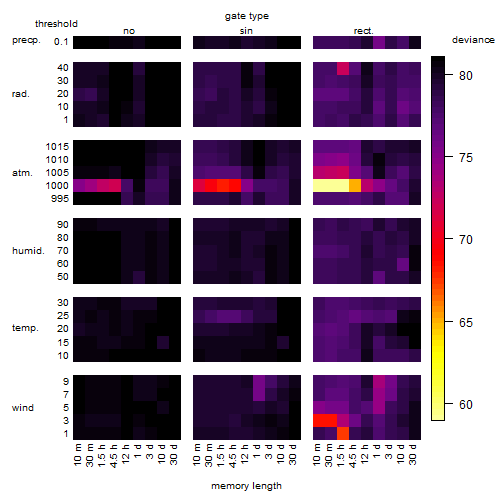
|
Histogram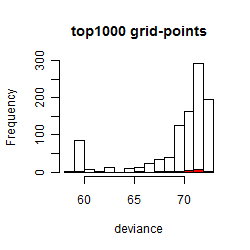 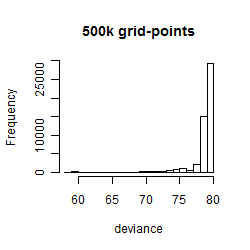
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 58.93 | atmosphere | 1000 | 90 | < th | dose dependent | rect. | 14 | 8 |
| 4 | 59.03 | atmosphere | 1000 | 90 | < th | dose dependent | rect. | 14 | 3 |
| 51 | 59.44 | atmosphere | 1000 | 90 | < th | dose independent | rect. | 14 | 1 |
| 72 | 59.82 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 14 | 8 |
| 117 | 65.10 | atmosphere | 1000 | 270 | < th | dose independent | rect. | 12 | 4 |
| 157 | 67.36 | wind | 1 | 90 | < th | dose independent | rect. | 14 | 1 |
| 166 | 67.62 | atmosphere | 1000 | 90 | < th | dose dependent | rect. | 14 | 15 |
| 167 | 67.64 | atmosphere | 1000 | 90 | < th | dose dependent | rect. | 14 | 17 |
| 193 | 68.09 | atmosphere | 1000 | 90 | < th | dose dependent | sin | 18 | NA |
| 204 | 68.30 | wind | 3 | 10 | < th | dose dependent | rect. | 16 | 1 |
| 234 | 69.11 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 12 | 10 |
| 252 | 69.19 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 12 | 5 |
| 361 | 70.07 | atmosphere | 1000 | 270 | < th | dose dependent | rect. | 11 | 19 |
| 452 | 70.65 | atmosphere | 1000 | 270 | < th | dose dependent | rect. | 23 | 17 |
| 464 | 70.70 | wind | 1 | 90 | < th | dose dependent | rect. | 14 | 1 |
| 509 | 70.98 | atmosphere | 1000 | 270 | < th | dose independent | sin | 21 | NA |
| 519 | 71.08 | atmosphere | 1000 | 90 | < th | dose independent | sin | 18 | NA |
| 616 | 71.42 | atmosphere | 1000 | 90 | < th | dose independent | rect. | 14 | 17 |
| 681 | 71.62 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 14 | 16 |
| 758 | 71.79 | atmosphere | 1000 | 270 | > th | dose independent | sin | 21 | NA |
| 794 | 71.93 | atmosphere | 1000 | 270 | < th | dose dependent | no | NA | NA |
| 800 | 71.97 | atmosphere | 1000 | 270 | < th | dose independent | rect. | 11 | 19 |
| 878 | 72.38 | radiation | 40 | 90 | > th | dose dependent | rect. | 14 | 1 |
| 960 | 72.63 | wind | 3 | 10 | < th | dose independent | rect. | 16 | 1 |