Os09g0474000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Eukaryotic transcription factor, DNA-binding domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os09g0474000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Eukaryotic transcription factor, DNA-binding domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

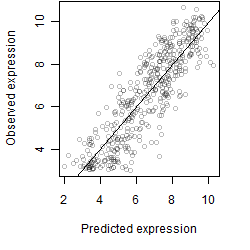
Dependence on each variable

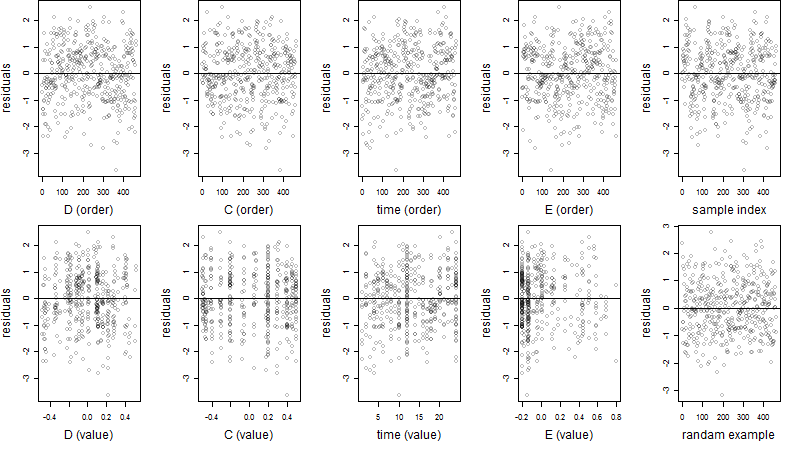
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 488.47 | 1.04 | 6.41 | -3.19 | 4.76 | 1.18 | 0.561 | 5.97 | 0.3 | 7.6 | 66.99 | 1473 | 10.1 | 2.15 |
| 525.41 | 1.07 | 6.3 | -2.56 | 4.64 | 2.04 | 0.963 | -- | 0.607 | 7.49 | 69 | 1793 | 9.13 | 1.12 |
| 487.67 | 1.03 | 6.42 | -3.19 | 4.74 | 1.19 | -- | 5.83 | 0.307 | 7.58 | 66.32 | 1481 | 10.3 | 2 |
| 522.47 | 1.06 | 6.29 | -2.44 | 4.62 | 2.4 | -- | -- | 0.589 | 7.37 | 68.56 | 1793 | 9.22 | 1.05 |
| 1641.69 | 1.89 | 6.56 | -2.39 | -- | 2.72 | -- | 3.57 | 0.499 | -- | 57.03 | 1564 | 10.9 | 0.1 |
| 608.75 | 1.16 | 6.35 | -2.67 | 4.79 | -- | 0.69 | -- | 0.333 | 7.7 | -- | -- | -- | -- |
| 610.09 | 1.16 | 6.35 | -2.66 | 4.79 | -- | -- | -- | 0.331 | 7.69 | -- | -- | -- | -- |
| 1592.12 | 1.86 | 6.32 | -1.84 | -- | 3.38 | -- | -- | 0.829 | -- | 6.246 | 3103 | 9.15 | 1.05 |
| 664.36 | 1.2 | 6.3 | -- | 4.57 | 2.87 | -- | -- | 0.595 | 7.21 | 66.45 | 1836 | 8.85 | 0.718 |
| 1844.69 | 2.01 | 6.47 | -2.59 | -- | -- | -- | -- | 0.402 | -- | -- | -- | -- | -- |
| 796.32 | 1.32 | 6.28 | -- | 4.77 | -- | -- | -- | 0.627 | 7.72 | -- | -- | -- | -- |
| 823.17 | 1.34 | 6.41 | -- | -- | 5.57 | -- | -- | 0.845 | -- | -54.25 | 3144 | 0.383 | 11.3 |
| 2022.32 | 2.1 | 6.4 | -- | -- | -- | -- | -- | 0.689 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 470.35 | temperature | 20 | 14400 | > th | dose dependent | rect. | 14 | 1 |
| 6 | 471.95 | wind | 9 | 43200 | < th | dose dependent | rect. | 4 | 2 |
| 13 | 473.05 | wind | 1 | 43200 | > th | dose dependent | rect. | 4 | 2 |
| 181 | 479.12 | temperature | 20 | 14400 | > th | dose dependent | sin | 23 | NA |
| 185 | 479.18 | wind | 1 | 43200 | > th | dose independent | rect. | 5 | 1 |
| 236 | 480.24 | temperature | 30 | 14400 | < th | dose dependent | rect. | 14 | 1 |
| 288 | 481.24 | temperature | 30 | 14400 | < th | dose dependent | rect. | 8 | 8 |
| 301 | 481.40 | wind | 1 | 43200 | > th | dose dependent | rect. | 23 | 7 |
| 338 | 481.92 | temperature | 30 | 14400 | < th | dose dependent | rect. | 8 | 3 |
| 346 | 482.03 | temperature | 25 | 14400 | > th | dose independent | rect. | 23 | 18 |
| 351 | 482.05 | wind | 1 | 43200 | > th | dose dependent | rect. | 1 | 5 |
| 400 | 482.42 | wind | 9 | 43200 | < th | dose dependent | rect. | 1 | 5 |
| 422 | 482.68 | wind | 9 | 43200 | < th | dose dependent | rect. | 23 | 7 |
| 439 | 482.82 | temperature | 25 | 14400 | < th | dose independent | rect. | 24 | 18 |
| 647 | 484.67 | temperature | 25 | 14400 | > th | dose independent | rect. | 10 | 18 |
| 697 | 485.02 | temperature | 30 | 14400 | < th | dose dependent | sin | 23 | NA |
| 699 | 485.04 | temperature | 25 | 14400 | > th | dose independent | rect. | 15 | 3 |
| 705 | 485.07 | temperature | 25 | 14400 | > th | dose independent | rect. | 11 | 10 |
| 728 | 485.23 | temperature | 25 | 14400 | > th | dose independent | rect. | 14 | 5 |
| 735 | 485.25 | temperature | 25 | 14400 | > th | dose independent | rect. | 13 | 7 |
| 756 | 485.37 | temperature | 25 | 14400 | > th | dose independent | sin | 3 | NA |
| 758 | 485.38 | temperature | 25 | 14400 | > th | dose independent | rect. | 5 | 13 |
| 834 | 485.84 | temperature | 25 | 14400 | < th | dose independent | rect. | 16 | 2 |
| 837 | 485.85 | temperature | 25 | 14400 | < th | dose independent | sin | 3 | NA |
| 918 | 486.24 | temperature | 25 | 14400 | > th | dose independent | sin | 20 | NA |
| 944 | 486.33 | temperature | 25 | 14400 | < th | dose independent | rect. | 10 | 18 |
| 973 | 486.49 | temperature | 25 | 14400 | < th | dose independent | rect. | 11 | 10 |