FiT-DB /
Search/
Help/
Sample detail
|
Os08g0484500
|
blastp(Os)/
blastp(At)/
coex///
RAP/
RiceXPro/
SALAD/
ATTED-II
|
|
Description : Similar to Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor (EC 2.5.1.54) (Phospho-2-keto-3-deoxyheptonate aldolase 1) (DAHP synthetase 1) (3-deoxy-D-arabino-heptulosonate 7-phosphate synthase 1).


|
log2(Expression) ~
Norm(μ, σ2)
μ = α
+ β1D
+ β2C
+ β3E
+ β4D*C
+ β5D*E
+ γ1N8
| par. |
|
value |
(S.E.) |
|
|
|
|
|
|
| α |
: |
14.2 |
(0.0185) |
R2 |
: |
0.445 |
|
|
|
| β1 |
: |
-- |
(--) |
R2dD |
: |
-- |
R2D |
: |
-- |
| β2 |
: |
0.736 |
(0.0465) |
R2dC |
: |
0.308 |
R2C |
: |
0.271 |
| β3 |
: |
-- |
(--) |
R2dE |
: |
-- |
R2E |
: |
-- |
| β4 |
: |
-- |
(--) |
R2dD*C |
: |
-- |
R2D*C |
: |
-- |
| β5 |
: |
-- |
(--) |
R2dD*E |
: |
-- |
R2D*E |
: |
-- |
| γ1 |
: |
-0.408 |
(0.0388) |
R2dN8 |
: |
0.174 |
R2N8 |
: |
0.137 |
| σ |
: |
0.348 |
|
|
|
|
|
|
|
| deviance |
: |
55.28 |
|
|
|
|
|
|
|
| __ |
|
|
|
|
|
|
|
|
|
|
_____
|
| C |
|
|
| peak time of C |
: |
20.6 |
| E |
|
|
| wheather |
: |
-- |
| threshold |
: |
-- |
| memory length |
: |
-- |
| response mode |
: |
-- |
| dose dependency |
: |
-- |
| G |
|
|
| type of G |
: |
-- |
| peak or start time of G |
: |
-- |
| open length of G |
: |
-- |
|

 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
| 24.89 |
0.234 |
14.1 |
1.17 |
0.75 |
-1.14 |
-0.0286 |
1.45 |
-0.0503 |
20.9 |
9.882 |
4162 |
-- |
-- |
| 28.08 |
0.247 |
14.1 |
0.956 |
0.697 |
-1.17 |
-0.215 |
-- |
-0.0643 |
21.1 |
9.08 |
4300 |
-- |
-- |
| 24.89 |
0.232 |
14.1 |
1.17 |
0.75 |
-1.14 |
-- |
1.45 |
-0.0503 |
20.9 |
9.883 |
4161 |
-- |
-- |
| 26.84 |
0.241 |
14.1 |
0.84 |
0.737 |
-1.2 |
-- |
-- |
-0.139 |
20.9 |
11.31 |
4305 |
-- |
-- |
| 55.82 |
0.348 |
14.1 |
1.16 |
-- |
-1.11 |
-- |
1.47 |
-0.0689 |
-- |
9.437 |
4393 |
-- |
-- |
| 53.13 |
0.342 |
14.2 |
0.281 |
0.737 |
-- |
-0.0942 |
-- |
-0.377 |
20.5 |
-- |
-- |
-- |
-- |
| 53.16 |
0.341 |
14.2 |
0.284 |
0.737 |
-- |
-- |
-- |
-0.377 |
20.5 |
-- |
-- |
-- |
-- |
| 57.77 |
0.354 |
14.1 |
0.836 |
-- |
-1.19 |
-- |
-- |
-0.148 |
-- |
11.49 |
4399 |
-- |
-- |
| 38.15 |
0.288 |
14.2 |
-- |
0.821 |
-0.919 |
-- |
-- |
-0.266 |
20.7 |
9.912 |
2027 |
-- |
-- |
| 83.63 |
0.427 |
14.2 |
0.281 |
-- |
-- |
-- |
-- |
-0.384 |
-- |
-- |
-- |
-- |
-- |
| 55.28 |
0.348 |
14.2 |
-- |
0.736 |
-- |
-- |
-- |
-0.408 |
20.6 |
-- |
-- |
-- |
-- |
| 70.84 |
0.392 |
14.1 |
-- |
-- |
-0.774 |
-- |
-- |
-0.291 |
-- |
6.874 |
2926 |
-- |
-- |
| 85.71 |
0.432 |
14.2 |
-- |
-- |
-- |
-- |
-- |
-0.415 |
-- |
-- |
-- |
-- |
-- |
Results of the grid search
Summarized heatmap of deviance
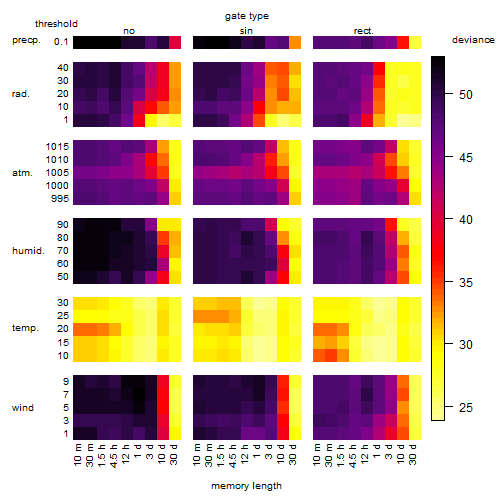
|
Histogram

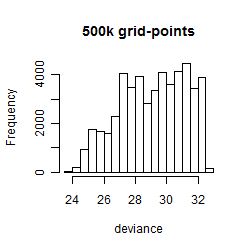
|
Local optima within top1000 grid-points
| rank |
deviance |
wheather |
threshold |
memory length |
response mode |
dose dependency |
type of G |
peak or start time of G |
open length of G |
| 1 |
23.84 |
temperature |
15 |
1440 |
> th |
dose dependent |
rect. |
18 |
14 |
| 9 |
23.93 |
temperature |
30 |
1440 |
< th |
dose dependent |
rect. |
18 |
14 |
| 28 |
24.00 |
temperature |
15 |
1440 |
> th |
dose dependent |
sin |
11 |
NA |
| 53 |
24.07 |
radiation |
1 |
14400 |
< th |
dose independent |
rect. |
18 |
13 |
| 67 |
24.11 |
temperature |
30 |
1440 |
< th |
dose dependent |
sin |
11 |
NA |
| 118 |
24.26 |
radiation |
1 |
14400 |
> th |
dose independent |
rect. |
18 |
13 |
| 146 |
24.34 |
temperature |
15 |
1440 |
> th |
dose dependent |
rect. |
1 |
7 |
| 150 |
24.35 |
temperature |
15 |
1440 |
> th |
dose dependent |
rect. |
3 |
5 |
| 167 |
24.37 |
temperature |
30 |
1440 |
< th |
dose dependent |
rect. |
1 |
7 |
| 173 |
24.38 |
temperature |
30 |
1440 |
< th |
dose dependent |
rect. |
3 |
5 |
| 277 |
24.57 |
temperature |
20 |
4320 |
> th |
dose dependent |
rect. |
15 |
4 |
| 316 |
24.61 |
temperature |
20 |
4320 |
> th |
dose dependent |
rect. |
13 |
9 |
| 357 |
24.65 |
temperature |
15 |
1440 |
> th |
dose dependent |
rect. |
21 |
2 |
| 391 |
24.67 |
temperature |
20 |
4320 |
> th |
dose dependent |
rect. |
17 |
1 |
| 439 |
24.71 |
temperature |
30 |
4320 |
< th |
dose dependent |
rect. |
21 |
1 |
| 483 |
24.73 |
radiation |
1 |
14400 |
< th |
dose dependent |
rect. |
4 |
3 |
| 606 |
24.80 |
temperature |
20 |
4320 |
> th |
dose dependent |
sin |
19 |
NA |
| 729 |
24.86 |
radiation |
1 |
14400 |
> th |
dose independent |
rect. |
17 |
19 |
| 825 |
24.90 |
radiation |
1 |
14400 |
< th |
dose independent |
rect. |
17 |
19 |
| 937 |
24.94 |
temperature |
10 |
4320 |
> th |
dose dependent |
no |
NA |
NA |
| 990 |
24.95 |
temperature |
10 |
4320 |
> th |
dose dependent |
rect. |
1 |
23 |
| 993 |
24.95 |
temperature |
10 |
4320 |
> th |
dose dependent |
rect. |
20 |
23 |



 __
__





