FiT-DB /
Search/
Help/
Sample detail
|
Os08g0424200
|
blastp(Os)/
blastp(At)/
coex///
RAP/
RiceXPro/
SALAD/
ATTED-II
|
|
Description : Similar to Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.4) (3-Methylcrotonyl-CoA carboxylase 2) (MCCase beta subunit) (3-methylcrotonyl-CoA:carbon dioxide ligase beta subunit).
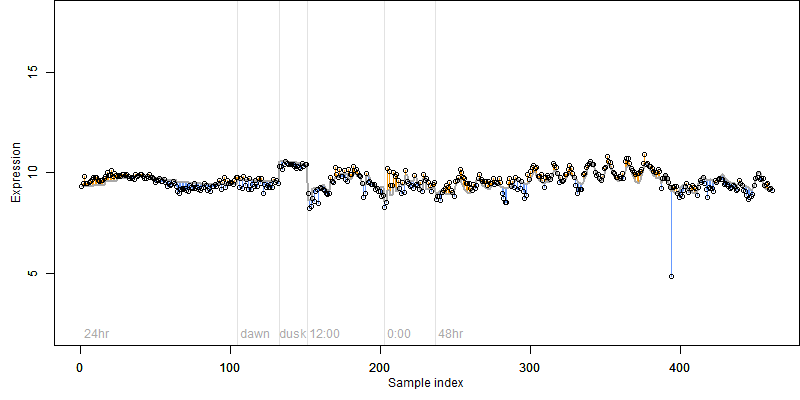

|
log2(Expression) ~
Norm(μ, σ2)
μ = α
+ β1D
+ β2C
+ β3E
+ β4D*C
+ β5D*E
+ γ1N8
| par. |
|
value |
(S.E.) |
|
|
|
|
|
|
| α |
: |
9.64 |
(0.0187) |
R2 |
: |
0.531 |
|
|
|
| β1 |
: |
-- |
(--) |
R2dD |
: |
-- |
R2D |
: |
-- |
| β2 |
: |
-- |
(--) |
R2dC |
: |
-- |
R2C |
: |
-- |
| β3 |
: |
1.99 |
(0.0875) |
R2dE |
: |
0.566 |
R2E |
: |
0.477 |
| β4 |
: |
-- |
(--) |
R2dD*C |
: |
-- |
R2D*C |
: |
-- |
| β5 |
: |
-- |
(--) |
R2dD*E |
: |
-- |
R2D*E |
: |
-- |
| γ1 |
: |
-0.249 |
(0.0404) |
R2dN8 |
: |
0.0539 |
R2N8 |
: |
-0.0345 |
| σ |
: |
0.349 |
|
|
|
|
|
|
|
| deviance |
: |
56.17 |
|
|
|
|
|
|
|
| __ |
|
|
|
|
|
|
|
|
|
|
_____
|
| C |
|
|
| peak time of C |
: |
-- |
| E |
|
|
| wheather |
: |
temperature |
| threshold |
: |
12.55 |
| memory length |
: |
339 |
| response mode |
: |
> th |
| dose dependency |
: |
dose dependent |
| G |
|
|
| type of G |
: |
no |
| peak or start time of G |
: |
-- |
| open length of G |
: |
-- |
|
 __
__
Dependence on each variable

Residual plot

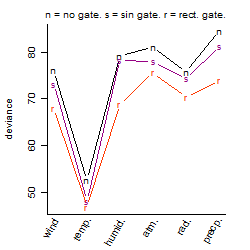
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
| 51.09 |
0.336 |
9.65 |
-0.357 |
0.287 |
2.17 |
0.386 |
-0.263 |
-0.322 |
10.5 |
9.433 |
460 |
-- |
-- |
| 51.13 |
0.333 |
9.65 |
-0.315 |
0.281 |
2.17 |
0.406 |
-- |
-0.311 |
10.4 |
9.461 |
454 |
-- |
-- |
| 51.67 |
0.335 |
9.66 |
-0.366 |
0.28 |
2.18 |
-- |
-0.411 |
-0.329 |
10.3 |
9.209 |
454 |
-- |
-- |
| 51.79 |
0.335 |
9.65 |
-0.298 |
0.276 |
2.17 |
-- |
-- |
-0.31 |
10.2 |
9.163 |
450 |
-- |
-- |
| 54.43 |
0.344 |
9.65 |
-0.299 |
-- |
2.08 |
-- |
-0.227 |
-0.297 |
-- |
10.05 |
346 |
-- |
-- |
| 103.85 |
0.478 |
9.59 |
0.181 |
0.513 |
-- |
0.549 |
-- |
0.00733 |
16.3 |
-- |
-- |
-- |
-- |
| 104.68 |
0.479 |
9.59 |
0.186 |
0.518 |
-- |
-- |
-- |
0.0103 |
16.4 |
-- |
-- |
-- |
-- |
| 54.46 |
0.344 |
9.65 |
-0.265 |
-- |
2.08 |
-- |
-- |
-0.289 |
-- |
13.26 |
346 |
-- |
-- |
| 53.97 |
0.342 |
9.65 |
-- |
0.251 |
2.08 |
-- |
-- |
-0.284 |
10.2 |
8.747 |
429 |
-- |
-- |
| 119.05 |
0.51 |
9.59 |
0.166 |
-- |
-- |
-- |
-- |
-0.00339 |
-- |
-- |
-- |
-- |
-- |
| 105.59 |
0.481 |
9.59 |
-- |
0.514 |
-- |
-- |
-- |
-0.0105 |
16.4 |
-- |
-- |
-- |
-- |
| 56.17 |
0.349 |
9.64 |
-- |
-- |
1.99 |
-- |
-- |
-0.249 |
-- |
12.55 |
339 |
-- |
-- |
| 119.78 |
0.511 |
9.59 |
-- |
-- |
-- |
-- |
-- |
-0.0218 |
-- |
-- |
-- |
-- |
-- |
Results of the grid search
Summarized heatmap of deviance

|
Histogram

|
Local optima within top1000 grid-points
| rank |
deviance |
wheather |
threshold |
memory length |
response mode |
dose dependency |
type of G |
peak or start time of G |
open length of G |
| 1 |
46.74 |
temperature |
15 |
720 |
> th |
dose dependent |
rect. |
23 |
17 |
| 14 |
48.00 |
temperature |
10 |
720 |
> th |
dose dependent |
sin |
4 |
NA |
| 21 |
48.30 |
temperature |
30 |
720 |
< th |
dose dependent |
rect. |
24 |
19 |
| 98 |
50.02 |
temperature |
30 |
720 |
< th |
dose dependent |
sin |
4 |
NA |
| 199 |
51.91 |
temperature |
10 |
720 |
> th |
dose dependent |
rect. |
12 |
23 |
| 237 |
52.47 |
temperature |
10 |
720 |
> th |
dose dependent |
no |
NA |
NA |
| 264 |
52.93 |
temperature |
30 |
720 |
< th |
dose dependent |
rect. |
17 |
21 |
| 268 |
52.97 |
temperature |
15 |
720 |
> th |
dose dependent |
rect. |
14 |
23 |
| 333 |
53.55 |
temperature |
10 |
720 |
> th |
dose dependent |
rect. |
10 |
23 |
| 386 |
53.97 |
temperature |
30 |
720 |
< th |
dose dependent |
no |
NA |
NA |
| 430 |
54.34 |
temperature |
15 |
720 |
> th |
dose dependent |
rect. |
8 |
23 |
| 487 |
54.98 |
temperature |
30 |
720 |
< th |
dose dependent |
rect. |
6 |
23 |
| 505 |
55.12 |
temperature |
30 |
720 |
< th |
dose dependent |
rect. |
4 |
23 |
| 515 |
55.20 |
temperature |
30 |
1440 |
< th |
dose dependent |
rect. |
17 |
1 |
| 695 |
56.49 |
temperature |
30 |
270 |
< th |
dose dependent |
sin |
23 |
NA |
| 725 |
56.65 |
temperature |
30 |
1440 |
< th |
dose dependent |
rect. |
9 |
13 |
| 754 |
56.76 |
temperature |
30 |
1440 |
< th |
dose dependent |
rect. |
15 |
5 |
| 756 |
56.77 |
temperature |
30 |
1440 |
< th |
dose dependent |
sin |
20 |
NA |
| 821 |
57.09 |
temperature |
30 |
1440 |
< th |
dose dependent |
rect. |
13 |
9 |
| 829 |
57.10 |
temperature |
10 |
1440 |
> th |
dose dependent |
rect. |
9 |
15 |
| 881 |
57.24 |
temperature |
10 |
1440 |
> th |
dose dependent |
sin |
19 |
NA |



 __
__





