Os08g0326000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Nucleoside phosphatase GDA1/CD39 family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os08g0326000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Nucleoside phosphatase GDA1/CD39 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
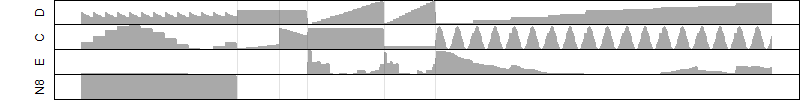
Dependence on each variable

Residual plot

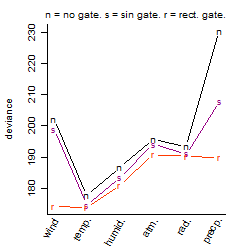
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 174.32 | 0.62 | 4.97 | 2.38 | 0.864 | 2.87 | -0.125 | 0.143 | 0.133 | 14.6 | 23.15 | 4096 | -- | -- |
| 174.28 | 0.615 | 4.96 | 2.41 | 0.875 | 2.86 | -0.12 | -- | 0.133 | 14.5 | 23.04 | 4154 | -- | -- |
| 174.28 | 0.615 | 4.97 | 2.38 | 0.869 | 2.93 | -- | 0.25 | 0.127 | 14.5 | 22.99 | 4145 | -- | -- |
| 174.34 | 0.615 | 4.96 | 2.41 | 0.874 | 2.87 | -- | -- | 0.134 | 14.5 | 23.03 | 4145 | -- | -- |
| 216.24 | 0.685 | 4.97 | 2.33 | -- | 3 | -- | 0.183 | 0.146 | -- | 23.84 | 3948 | -- | -- |
| 300.44 | 0.813 | 5.12 | 0.905 | 0.986 | -- | -0.279 | -- | -0.58 | 14.6 | -- | -- | -- | -- |
| 300.71 | 0.812 | 5.12 | 0.898 | 0.985 | -- | -- | -- | -0.581 | 14.6 | -- | -- | -- | -- |
| 216.09 | 0.685 | 4.96 | 2.36 | -- | 2.94 | -- | -- | 0.163 | -- | 24.05 | 3945 | -- | -- |
| 254.25 | 0.743 | 5.07 | -- | 0.845 | 2.16 | -- | -- | -0.337 | 14.7 | 24 | 1256 | -- | -- |
| 357.52 | 0.884 | 5.13 | 0.856 | -- | -- | -- | -- | -0.606 | -- | -- | -- | -- | -- |
| 321.97 | 0.839 | 5.15 | -- | 0.967 | -- | -- | -- | -0.681 | 14.6 | -- | -- | -- | -- |
| 287.98 | 0.79 | 5.05 | -- | -- | 2.25 | -- | -- | -0.268 | -- | 26.33 | 1108 | -- | -- |
| 376.86 | 0.906 | 5.15 | -- | -- | -- | -- | -- | -0.701 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 173.91 | temperature | 25 | 4320 | < th | dose dependent | rect. | 2 | 14 |
| 7 | 174.22 | temperature | 25 | 1440 | < th | dose dependent | rect. | 5 | 6 |
| 8 | 174.29 | wind | 3 | 43200 | < th | dose dependent | rect. | 23 | 1 |
| 16 | 174.63 | temperature | 25 | 4320 | < th | dose dependent | sin | 3 | NA |
| 171 | 177.67 | temperature | 25 | 4320 | < th | dose dependent | no | NA | NA |
| 172 | 177.67 | temperature | 25 | 4320 | < th | dose dependent | rect. | 5 | 23 |
| 184 | 177.80 | temperature | 25 | 4320 | < th | dose dependent | rect. | 8 | 23 |
| 205 | 177.98 | temperature | 25 | 4320 | < th | dose dependent | rect. | 13 | 23 |
| 239 | 178.37 | temperature | 20 | 1440 | > th | dose independent | rect. | 2 | 12 |
| 292 | 178.84 | temperature | 20 | 1440 | < th | dose independent | rect. | 2 | 12 |
| 376 | 179.30 | temperature | 20 | 1440 | > th | dose independent | rect. | 2 | 16 |
| 440 | 179.59 | temperature | 20 | 4320 | < th | dose dependent | rect. | 3 | 1 |
| 496 | 179.88 | temperature | 20 | 1440 | < th | dose independent | rect. | 2 | 16 |
| 559 | 180.08 | temperature | 20 | 1440 | > th | dose independent | sin | 4 | NA |
| 571 | 180.11 | temperature | 10 | 4320 | > th | dose dependent | rect. | 1 | 11 |
| 613 | 180.23 | temperature | 10 | 4320 | > th | dose dependent | rect. | 23 | 13 |
| 644 | 180.36 | temperature | 20 | 1440 | < th | dose independent | sin | 4 | NA |
| 645 | 180.36 | temperature | 20 | 4320 | < th | dose dependent | rect. | 22 | 2 |
| 718 | 180.61 | wind | 1 | 43200 | > th | dose dependent | rect. | 23 | 1 |
| 795 | 180.80 | temperature | 10 | 4320 | > th | dose dependent | sin | 5 | NA |
| 866 | 180.99 | humidity | 50 | 43200 | > th | dose dependent | rect. | 21 | 1 |