Os08g0219100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os08g0219100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

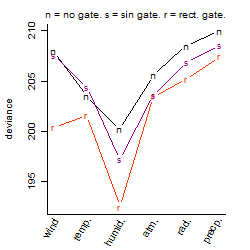
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 194.43 | 0.655 | 2.96 | -0.156 | 0.0473 | 5.8 | 0.943 | -10.3 | 0.0191 | 0.969 | 48.26 | 666 | -- | -- |
| 202.66 | 0.663 | 2.91 | -0.0512 | 0.148 | 1.96 | 0.648 | -- | 0.0127 | 22.8 | 48.04 | 1134 | -- | -- |
| 197.08 | 0.654 | 2.96 | -0.21 | 0.0465 | 6.48 | -- | -11.4 | 0.00771 | 1.34 | 47.98 | 671 | -- | -- |
| 204.37 | 0.666 | 2.91 | -0.0904 | 0.0702 | 2.1 | -- | -- | -0.00505 | 22.1 | 48 | 1135 | -- | -- |
| 197.2 | 0.654 | 2.97 | -0.192 | -- | 6.52 | -- | -11.5 | 0.0105 | -- | 47.97 | 671 | -- | -- |
| 214.07 | 0.686 | 2.91 | 0.119 | 0.121 | -- | 0.97 | -- | -0.0224 | 23.3 | -- | -- | -- | -- |
| 218.11 | 0.692 | 2.92 | 0.0799 | 0.117 | -- | -- | -- | -0.0261 | 23.4 | -- | -- | -- | -- |
| 202.56 | 0.663 | 2.91 | -0.0938 | -- | 2.22 | -- | -- | 0.00276 | -- | 48.87 | 714 | -- | -- |
| 204.59 | 0.666 | 2.91 | -- | 0.0671 | 2.03 | -- | -- | 0.00253 | 23.2 | 48 | 1135 | -- | -- |
| 218.96 | 0.691 | 2.91 | 0.0829 | -- | -- | -- | -- | -0.0249 | -- | -- | -- | -- | -- |
| 218.28 | 0.691 | 2.92 | -- | 0.118 | -- | -- | -- | -0.035 | 23.4 | -- | -- | -- | -- |
| 202.71 | 0.663 | 2.91 | -- | -- | 2.2 | -- | -- | 0.00615 | -- | 48.56 | 712 | -- | -- |
| 219.15 | 0.691 | 2.91 | -- | -- | -- | -- | -- | -0.0341 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 192.47 | humidity | 50 | 720 | < th | dose independent | rect. | 12 | 1 |
| 3 | 193.74 | humidity | 50 | 720 | < th | dose dependent | rect. | 13 | 3 |
| 91 | 197.21 | humidity | 50 | 720 | < th | dose dependent | sin | 6 | NA |
| 227 | 200.17 | humidity | 50 | 720 | < th | dose dependent | no | NA | NA |
| 387 | 200.40 | wind | 1 | 90 | < th | dose independent | rect. | 2 | 1 |
| 388 | 200.42 | humidity | 50 | 720 | < th | dose independent | sin | 7 | NA |
| 413 | 200.89 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 11 |
| 416 | 200.95 | humidity | 50 | 4320 | < th | dose dependent | rect. | 17 | 1 |
| 432 | 200.99 | humidity | 50 | 4320 | < th | dose independent | rect. | 17 | 1 |
| 470 | 201.06 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 1 |
| 471 | 201.11 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 16 |
| 478 | 201.28 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 13 |
| 480 | 201.32 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 5 |
| 559 | 201.60 | temperature | 25 | 10 | < th | dose dependent | rect. | 9 | 19 |
| 594 | 201.86 | humidity | 70 | 1440 | < th | dose dependent | rect. | 19 | 1 |
| 684 | 202.04 | wind | 1 | 270 | < th | dose dependent | rect. | 21 | 1 |
| 690 | 202.11 | humidity | 70 | 720 | < th | dose independent | rect. | 19 | 1 |
| 809 | 202.35 | temperature | 25 | 10 | < th | dose dependent | rect. | 6 | 22 |
| 865 | 202.54 | humidity | 50 | 1440 | > th | dose independent | rect. | 12 | 1 |
| 872 | 202.64 | humidity | 50 | 1440 | > th | dose independent | rect. | 5 | 10 |
| 888 | 202.73 | humidity | 50 | 1440 | > th | dose independent | rect. | 18 | 21 |
| 889 | 202.74 | temperature | 25 | 10 | < th | dose dependent | rect. | 19 | 21 |
| 897 | 202.77 | humidity | 50 | 1440 | > th | dose independent | rect. | 23 | 16 |
| 898 | 202.77 | humidity | 50 | 1440 | > th | dose independent | rect. | 20 | 19 |
| 933 | 202.85 | humidity | 50 | 1440 | > th | dose independent | rect. | 10 | 5 |
| 950 | 202.86 | temperature | 25 | 10 | < th | dose dependent | rect. | 20 | 18 |
| 991 | 202.95 | temperature | 20 | 10 | > th | dose independent | rect. | 4 | 23 |