Os07g0667400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : SAM (and some other nucleotide) binding motif domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os07g0667400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : SAM (and some other nucleotide) binding motif domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

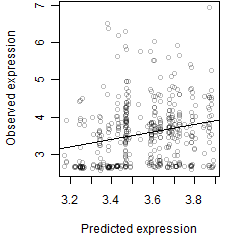 __
__
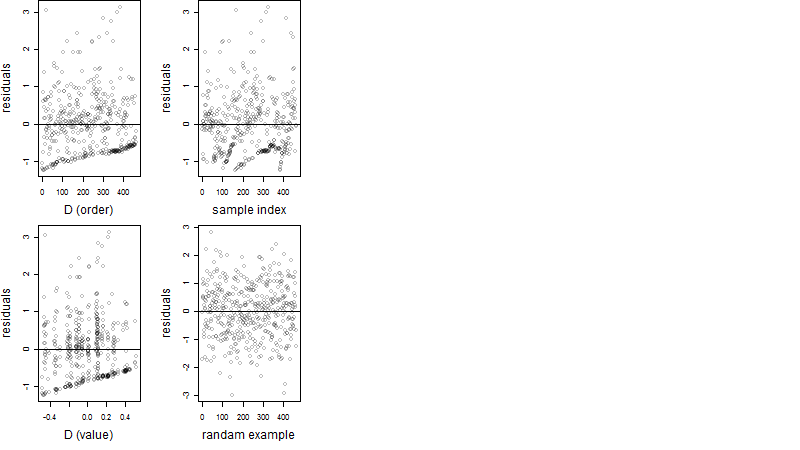
Dependence on each variable

Residual plot
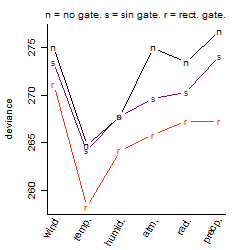
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 263.05 | 0.762 | 3.59 | -1.26 | 0.389 | 1.52 | 0.047 | -4.06 | -0.0456 | 4.98 | 26.64 | 200 | -- | -- |
| 265.57 | 0.759 | 3.55 | -0.903 | 0.406 | 1.15 | 0.482 | -- | -0.0127 | 5.02 | 25.9 | 248 | -- | -- |
| 263.05 | 0.755 | 3.59 | -1.26 | 0.39 | 1.52 | -- | -4.08 | -0.0474 | 5.02 | 26.61 | 201 | -- | -- |
| 265.87 | 0.759 | 3.54 | -0.875 | 0.386 | 1.09 | -- | -- | 0.017 | 5.39 | 26 | 251 | -- | -- |
| 267.89 | 0.762 | 3.58 | -1.14 | -- | 1.27 | -- | -3.95 | 0.00569 | -- | 27.42 | 191 | -- | -- |
| 285.18 | 0.792 | 3.54 | -0.696 | 0.0772 | -- | -0.502 | -- | 0.0511 | 11.1 | -- | -- | -- | -- |
| 286.06 | 0.792 | 3.54 | -0.714 | 0.0988 | -- | -- | -- | 0.0509 | 13.7 | -- | -- | -- | -- |
| 270.47 | 0.766 | 3.54 | -0.82 | -- | 0.858 | -- | -- | 0.0495 | -- | 27.01 | 201 | -- | -- |
| 285.46 | 0.787 | 3.52 | -- | 0.259 | 0.891 | -- | -- | 0.126 | 5.47 | 26.59 | 248 | -- | -- |
| 286.65 | 0.791 | 3.55 | -0.718 | -- | -- | -- | -- | 0.0486 | -- | -- | -- | -- | -- |
| 299.49 | 0.81 | 3.53 | -- | 0.112 | -- | -- | -- | 0.13 | 13.9 | -- | -- | -- | -- |
| 287.87 | 0.79 | 3.52 | -- | -- | 0.757 | -- | -- | 0.146 | -- | 27.4 | 217 | -- | -- |
| 300.25 | 0.809 | 3.53 | -- | -- | -- | -- | -- | 0.128 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance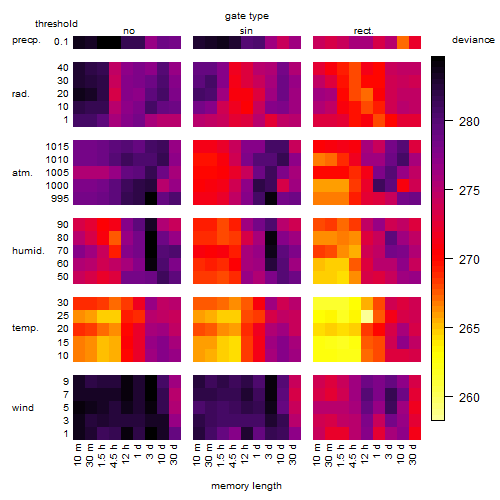
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 258.27 | temperature | 25 | 720 | > th | dose dependent | rect. | 6 | 2 |
| 5 | 261.39 | temperature | 30 | 30 | < th | dose dependent | rect. | 10 | 15 |
| 10 | 261.78 | temperature | 25 | 90 | > th | dose dependent | rect. | 11 | 23 |
| 12 | 261.87 | temperature | 15 | 90 | > th | dose dependent | rect. | 11 | 23 |
| 16 | 262.11 | temperature | 15 | 30 | > th | dose dependent | rect. | 21 | 23 |
| 21 | 262.25 | temperature | 10 | 270 | > th | dose dependent | rect. | 21 | 22 |
| 24 | 262.29 | temperature | 30 | 10 | < th | dose independent | rect. | 10 | 15 |
| 26 | 262.48 | temperature | 30 | 90 | < th | dose dependent | rect. | 3 | 21 |
| 37 | 262.91 | temperature | 25 | 10 | > th | dose dependent | rect. | 12 | 22 |
| 39 | 263.06 | temperature | 25 | 720 | > th | dose dependent | rect. | 22 | 11 |
| 40 | 263.06 | temperature | 15 | 10 | > th | dose dependent | rect. | 12 | 23 |
| 42 | 263.16 | temperature | 30 | 270 | < th | dose independent | rect. | 13 | 23 |
| 43 | 263.18 | temperature | 30 | 10 | < th | dose dependent | rect. | 4 | 21 |
| 57 | 263.52 | temperature | 15 | 90 | > th | dose dependent | rect. | 1 | 18 |
| 59 | 263.54 | temperature | 25 | 270 | > th | dose dependent | rect. | 5 | 13 |
| 66 | 263.64 | temperature | 25 | 90 | > th | dose dependent | rect. | 21 | 23 |
| 69 | 263.68 | temperature | 30 | 270 | < th | dose independent | rect. | 7 | 14 |
| 74 | 263.70 | temperature | 25 | 270 | > th | dose dependent | rect. | 23 | 19 |
| 78 | 263.72 | temperature | 25 | 270 | > th | dose dependent | rect. | 21 | 21 |
| 101 | 263.90 | temperature | 15 | 10 | > th | dose dependent | rect. | 1 | 19 |
| 130 | 264.15 | humidity | 50 | 90 | > th | dose dependent | rect. | 10 | 14 |
| 136 | 264.21 | temperature | 10 | 270 | > th | dose dependent | sin | 21 | NA |
| 157 | 264.34 | temperature | 15 | 90 | > th | dose dependent | rect. | 23 | 23 |
| 172 | 264.41 | temperature | 25 | 90 | > th | dose dependent | rect. | 2 | 22 |
| 187 | 264.48 | temperature | 10 | 90 | > th | dose dependent | sin | 19 | NA |
| 218 | 264.56 | temperature | 25 | 270 | > th | dose dependent | sin | 23 | NA |
| 254 | 264.68 | temperature | 15 | 270 | > th | dose dependent | rect. | 8 | 23 |
| 257 | 264.70 | temperature | 25 | 270 | > th | dose dependent | no | NA | NA |
| 308 | 264.84 | temperature | 25 | 90 | > th | dose dependent | sin | 20 | NA |
| 366 | 265.06 | temperature | 10 | 90 | > th | dose dependent | no | NA | NA |
| 378 | 265.09 | temperature | 25 | 90 | > th | dose dependent | rect. | 15 | 23 |
| 417 | 265.30 | temperature | 30 | 270 | < th | dose independent | rect. | 2 | 23 |
| 450 | 265.45 | temperature | 15 | 270 | > th | dose dependent | rect. | 5 | 13 |
| 463 | 265.48 | temperature | 30 | 720 | < th | dose independent | rect. | 17 | 21 |
| 480 | 265.54 | humidity | 70 | 270 | > th | dose independent | rect. | 16 | 20 |
| 497 | 265.58 | temperature | 25 | 30 | > th | dose dependent | rect. | 12 | 8 |
| 555 | 265.80 | atmosphere | 1000 | 10 | > th | dose dependent | rect. | 19 | 6 |
| 573 | 265.85 | atmosphere | 1000 | 90 | > th | dose dependent | rect. | 18 | 6 |
| 610 | 265.95 | temperature | 30 | 720 | < th | dose independent | rect. | 2 | 12 |
| 637 | 266.08 | temperature | 30 | 720 | < th | dose independent | rect. | 9 | 5 |
| 660 | 266.13 | temperature | 20 | 30 | < th | dose dependent | rect. | 1 | 1 |
| 665 | 266.14 | humidity | 50 | 270 | > th | dose dependent | rect. | 7 | 14 |
| 677 | 266.18 | humidity | 70 | 90 | > th | dose independent | rect. | 10 | 14 |
| 682 | 266.18 | temperature | 15 | 270 | > th | dose dependent | rect. | 5 | 23 |
| 687 | 266.20 | temperature | 30 | 720 | < th | dose independent | rect. | 3 | 9 |
| 725 | 266.29 | humidity | 80 | 270 | < th | dose dependent | rect. | 12 | 23 |
| 726 | 266.30 | temperature | 30 | 270 | < th | dose independent | sin | 22 | NA |
| 793 | 266.46 | temperature | 30 | 720 | < th | dose independent | rect. | 21 | 17 |
| 812 | 266.49 | temperature | 25 | 90 | > th | dose dependent | rect. | 11 | 13 |
| 827 | 266.52 | temperature | 30 | 720 | < th | dose independent | rect. | 2 | 19 |
| 828 | 266.52 | temperature | 30 | 270 | < th | dose dependent | sin | 22 | NA |
| 832 | 266.53 | temperature | 30 | 270 | > th | dose independent | rect. | 8 | 10 |
| 869 | 266.56 | temperature | 30 | 720 | < th | dose independent | rect. | 9 | 20 |
| 870 | 266.56 | temperature | 25 | 90 | > th | dose dependent | rect. | 11 | 11 |
| 876 | 266.57 | temperature | 30 | 90 | < th | dose dependent | rect. | 9 | 23 |
| 882 | 266.59 | temperature | 30 | 270 | < th | dose independent | rect. | 6 | 23 |
| 895 | 266.63 | atmosphere | 1010 | 10 | < th | dose dependent | rect. | 16 | 15 |
| 977 | 266.82 | temperature | 30 | 720 | < th | dose independent | rect. | 24 | 21 |
| 982 | 266.82 | temperature | 25 | 90 | > th | dose dependent | rect. | 11 | 17 |
| 986 | 266.83 | temperature | 20 | 90 | < th | dose independent | rect. | 24 | 1 |