Os07g0633000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.
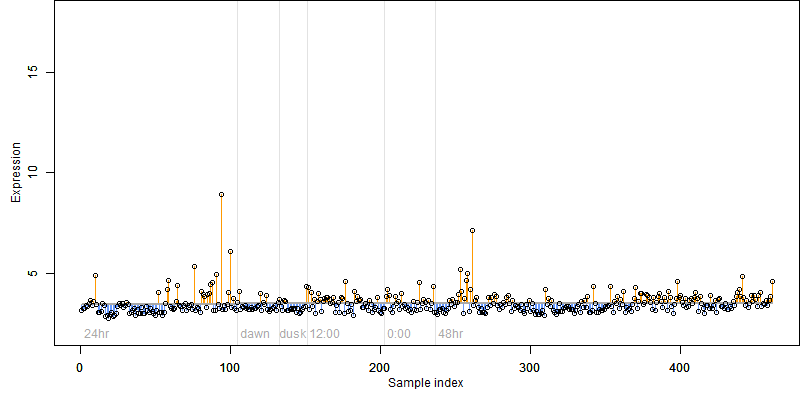

FiT-DB / Search/ Help/ Sample detail
|
Os07g0633000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

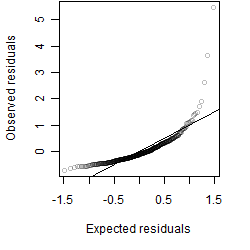
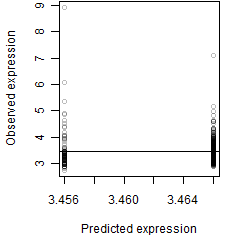
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 114.12 | 0.502 | 3.47 | -0.0591 | 0.276 | -0.403 | -0.971 | 2.37 | 0.0244 | 5.79 | 62 | 325 | -- | -- |
| 118.6 | 0.507 | 3.46 | 0.000845 | 0.282 | -0.172 | -0.571 | -- | 0.00262 | 6.05 | 64 | 348 | -- | -- |
| 116.12 | 0.502 | 3.47 | -0.069 | 0.3 | -0.441 | -- | 2.04 | 0.0293 | 5.97 | 62 | 327 | -- | -- |
| 119.33 | 0.509 | 3.45 | -0.0162 | 0.294 | -0.218 | -- | -- | 0.00906 | 6.25 | 62.01 | 380 | -- | -- |
| 119.72 | 0.51 | 3.48 | -0.0369 | -- | -0.242 | -- | 1.88 | 0.0164 | -- | 62.17 | 321 | -- | -- |
| 119.15 | 0.512 | 3.46 | 0.0115 | 0.229 | -- | -0.678 | -- | -0.016 | 6.46 | -- | -- | -- | -- |
| 120.3 | 0.514 | 3.46 | 0.0024 | 0.226 | -- | -- | -- | -0.0138 | 6.5 | -- | -- | -- | -- |
| 122.61 | 0.518 | 3.46 | 0.00729 | -- | -0.107 | -- | -- | 0.00118 | -- | 58 | 267 | -- | -- |
| 119.34 | 0.509 | 3.45 | -- | 0.294 | -0.215 | -- | -- | 0.0107 | 6.25 | 62.03 | 380 | -- | -- |
| 122.95 | 0.518 | 3.47 | 0.00803 | -- | -- | -- | -- | -0.00914 | -- | -- | -- | -- | -- |
| 120.3 | 0.513 | 3.46 | -- | 0.226 | -- | -- | -- | -0.0141 | 6.5 | -- | -- | -- | -- |
| 122.6 | 0.516 | 3.46 | -- | -- | 0.274 | -- | -- | -0.0207 | -- | 50 | 189 | -- | -- |
| 122.95 | 0.518 | 3.47 | -- | -- | -- | -- | -- | -0.01 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram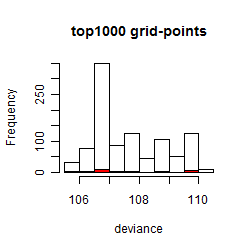 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 105.73 | humidity | 60 | 720 | < th | dose dependent | rect. | 19 | 15 |
| 5 | 105.97 | humidity | 60 | 270 | < th | dose dependent | rect. | 9 | 1 |
| 33 | 106.06 | humidity | 60 | 90 | < th | dose independent | rect. | 18 | 15 |
| 40 | 106.43 | humidity | 60 | 270 | < th | dose independent | rect. | 8 | 1 |
| 165 | 106.56 | humidity | 60 | 30 | < th | dose independent | rect. | 16 | 18 |
| 168 | 106.56 | humidity | 60 | 10 | < th | dose dependent | rect. | 16 | 18 |
| 207 | 106.64 | humidity | 60 | 10 | < th | dose independent | rect. | 10 | 1 |
| 209 | 106.64 | humidity | 60 | 1440 | > th | dose independent | rect. | 22 | 11 |
| 210 | 106.64 | humidity | 60 | 10 | < th | dose dependent | rect. | 10 | 1 |
| 277 | 106.69 | wind | 1 | 10 | < th | dose dependent | rect. | 8 | 2 |
| 336 | 106.72 | wind | 1 | 10 | < th | dose dependent | rect. | 9 | 5 |
| 339 | 106.74 | humidity | 60 | 1440 | > th | dose independent | rect. | 6 | 3 |
| 405 | 106.85 | wind | 1 | 10 | < th | dose independent | rect. | 8 | 2 |
| 461 | 107.07 | humidity | 60 | 1440 | > th | dose independent | rect. | 4 | 5 |
| 465 | 107.08 | wind | 1 | 10 | < th | dose independent | rect. | 9 | 5 |
| 482 | 107.19 | humidity | 60 | 1440 | > th | dose independent | rect. | 8 | 1 |
| 559 | 107.62 | radiation | 40 | 10 | > th | dose dependent | rect. | 9 | 1 |
| 715 | 108.52 | radiation | 40 | 10 | > th | dose dependent | sin | 11 | NA |
| 786 | 108.86 | humidity | 60 | 10 | < th | dose independent | sin | 10 | NA |
| 871 | 109.55 | radiation | 40 | 10 | > th | dose independent | rect. | 9 | 1 |
| 915 | 109.76 | humidity | 60 | 1440 | > th | dose independent | rect. | 19 | 15 |
| 965 | 109.89 | radiation | 20 | 270 | > th | dose dependent | rect. | 6 | 1 |
| 985 | 109.98 | humidity | 60 | 10 | > th | dose independent | sin | 10 | NA |