Os07g0411300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Type III polyketide synthase family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os07g0411300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Type III polyketide synthase family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 77.96 | 0.415 | 2.91 | -0.15 | 0.0834 | 0.0978 | 0.0486 | -1.37 | 0.0296 | 6.65 | 1.777 | 1472 | -- | -- |
| 79.49 | 0.415 | 2.93 | -0.181 | 0.0754 | -0.127 | 0.197 | -- | 0.0398 | 6.07 | 5.999 | 2446 | -- | -- |
| 77.96 | 0.411 | 2.91 | -0.15 | 0.0836 | 0.0977 | -- | -1.38 | 0.0295 | 6.65 | 1.778 | 1474 | -- | -- |
| 79.46 | 0.415 | 2.93 | -0.188 | 0.0721 | -0.143 | -- | -- | 0.0349 | 6.04 | 6.4 | 4545 | -- | -- |
| 78.32 | 0.412 | 2.91 | -0.151 | -- | 0.0779 | -- | -1.32 | 0.0349 | -- | 1.8 | 1474 | -- | -- |
| 79.81 | 0.419 | 2.94 | -0.163 | 0.075 | -- | 0.165 | -- | 0.0385 | 6.25 | -- | -- | -- | -- |
| 79.88 | 0.419 | 2.94 | -0.161 | 0.076 | -- | -- | -- | 0.0379 | 6.04 | -- | -- | -- | -- |
| 79.59 | 0.415 | 2.93 | -0.219 | -- | -0.137 | -- | -- | 0.0538 | -- | 7.397 | 5637 | -- | -- |
| 80.48 | 0.418 | 2.93 | -- | 0.0627 | -0.0617 | -- | -- | 0.0491 | 6.05 | 6.8 | 4543 | -- | -- |
| 80.18 | 0.418 | 2.94 | -0.159 | -- | -- | -- | -- | 0.0396 | -- | -- | -- | -- | -- |
| 80.56 | 0.42 | 2.93 | -- | 0.0736 | -- | -- | -- | 0.0558 | 6.15 | -- | -- | -- | -- |
| 80.55 | 0.419 | 2.93 | -- | -- | 0.258 | -- | -- | 0.0616 | -- | 8.7 | 5639 | -- | -- |
| 80.84 | 0.42 | 2.93 | -- | -- | -- | -- | -- | 0.0572 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 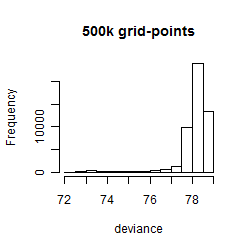
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 72.47 | humidity | 60 | 1440 | > th | dose independent | rect. | 23 | 10 |
| 2 | 72.72 | humidity | 60 | 1440 | > th | dose independent | rect. | 6 | 3 |
| 4 | 72.75 | humidity | 60 | 1440 | > th | dose independent | rect. | 8 | 1 |
| 5 | 72.81 | humidity | 60 | 1440 | < th | dose dependent | rect. | 8 | 1 |
| 31 | 72.82 | humidity | 60 | 1440 | < th | dose independent | rect. | 8 | 1 |
| 45 | 72.87 | humidity | 60 | 270 | < th | dose independent | rect. | 8 | 1 |
| 72 | 72.87 | humidity | 60 | 270 | < th | dose dependent | rect. | 8 | 1 |
| 114 | 73.22 | wind | 1 | 10 | < th | dose dependent | rect. | 9 | 1 |
| 116 | 73.22 | humidity | 60 | 90 | < th | dose dependent | rect. | 18 | 16 |
| 118 | 73.23 | humidity | 60 | 90 | < th | dose independent | rect. | 18 | 15 |
| 120 | 73.24 | humidity | 60 | 90 | < th | dose independent | rect. | 17 | 17 |
| 144 | 73.31 | humidity | 60 | 10 | < th | dose independent | rect. | 9 | 1 |
| 204 | 73.31 | humidity | 60 | 10 | < th | dose dependent | rect. | 9 | 1 |
| 300 | 73.33 | wind | 1 | 10 | < th | dose independent | rect. | 9 | 1 |
| 305 | 73.35 | radiation | 40 | 10 | > th | dose dependent | rect. | 9 | 1 |
| 328 | 73.40 | humidity | 60 | 1440 | > th | dose independent | rect. | 3 | 6 |
| 604 | 74.07 | radiation | 40 | 10 | > th | dose independent | rect. | 14 | 20 |
| 616 | 74.11 | radiation | 40 | 10 | > th | dose independent | rect. | 9 | 1 |
| 819 | 74.67 | radiation | 40 | 10 | > th | dose dependent | sin | 12 | NA |