Os07g0291100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Transport protein particle (TRAPP) component, Bet3 family protein.
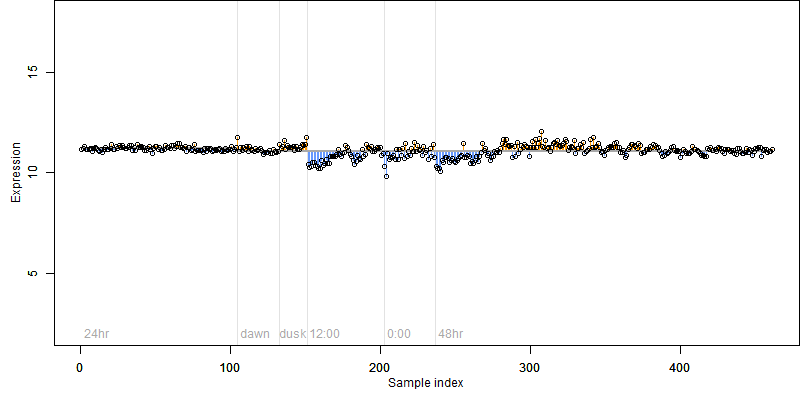

FiT-DB / Search/ Help/ Sample detail
|
Os07g0291100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Transport protein particle (TRAPP) component, Bet3 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
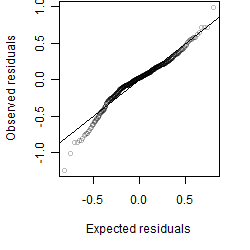
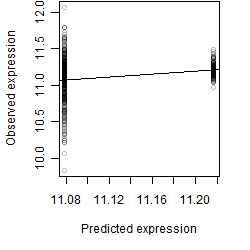 __
__
Dependence on each variable

Residual plot

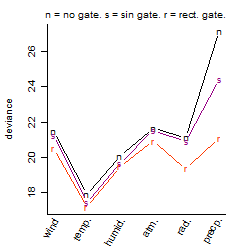
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17.79 | 0.198 | 11.1 | -0.213 | 0.185 | 0.857 | -0.426 | -2.4 | -0.0554 | 3.9 | 11.71 | 604 | -- | -- |
| 21.93 | 0.218 | 11.1 | 0.21 | 0.181 | 0.773 | -0.33 | -- | 0.0629 | 2.97 | 4.254 | 612 | -- | -- |
| 18.16 | 0.198 | 11.1 | -0.219 | 0.101 | 0.799 | -- | -2.29 | -0.0525 | 3.26 | 13.95 | 783 | -- | -- |
| 22.26 | 0.22 | 11.1 | 0.208 | 0.19 | 0.812 | -- | -- | 0.0581 | 3.63 | 13.7 | 604 | -- | -- |
| 18.37 | 0.2 | 11.2 | -0.264 | -- | 0.799 | -- | -2.32 | -0.0632 | -- | 14.01 | 948 | -- | -- |
| 29.56 | 0.255 | 11.1 | 0.388 | 0.192 | -- | -0.359 | -- | 0.18 | 23.6 | -- | -- | -- | -- |
| 30.11 | 0.257 | 11.1 | 0.403 | 0.194 | -- | -- | -- | 0.181 | 23.3 | -- | -- | -- | -- |
| 22.83 | 0.223 | 11.1 | 0.196 | -- | 0.778 | -- | -- | 0.0593 | -- | 12.32 | 888 | -- | -- |
| 23.28 | 0.225 | 11.1 | -- | 0.204 | 0.906 | -- | -- | 0.0235 | 4.05 | 13.06 | 614 | -- | -- |
| 32.47 | 0.266 | 11.1 | 0.408 | -- | -- | -- | -- | 0.183 | -- | -- | -- | -- | -- |
| 34.4 | 0.274 | 11.1 | -- | 0.199 | -- | -- | -- | 0.136 | 23.4 | -- | -- | -- | -- |
| 23.72 | 0.227 | 11.1 | -- | -- | 0.854 | -- | -- | 0.0265 | -- | 7.484 | 916 | -- | -- |
| 36.87 | 0.283 | 11.1 | -- | -- | -- | -- | -- | 0.138 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance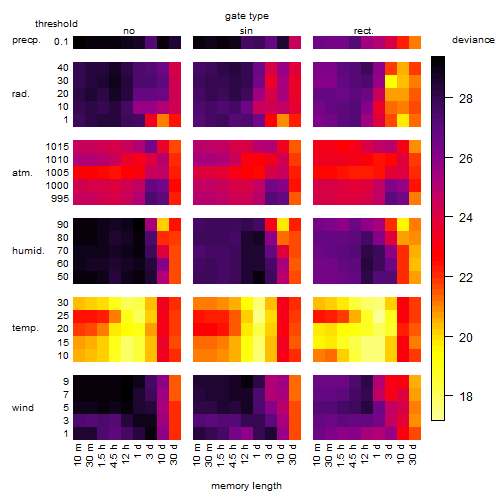
|
Histogram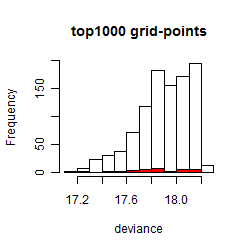 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 17.16 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 3 | 17.27 | temperature | 25 | 1440 | < th | dose dependent | rect. | 19 | 11 |
| 36 | 17.43 | temperature | 25 | 1440 | < th | dose dependent | sin | 11 | NA |
| 75 | 17.57 | temperature | 10 | 1440 | > th | dose dependent | rect. | 19 | 11 |
| 139 | 17.67 | temperature | 10 | 720 | > th | dose dependent | rect. | 1 | 23 |
| 148 | 17.68 | temperature | 15 | 720 | > th | dose dependent | rect. | 19 | 22 |
| 155 | 17.70 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 3 |
| 169 | 17.71 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 5 |
| 191 | 17.73 | temperature | 10 | 1440 | > th | dose dependent | sin | 11 | NA |
| 192 | 17.73 | temperature | 10 | 1440 | > th | dose dependent | rect. | 21 | 1 |
| 244 | 17.78 | temperature | 10 | 1440 | > th | dose dependent | rect. | 3 | 3 |
| 245 | 17.78 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 4 |
| 317 | 17.82 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 9 |
| 344 | 17.83 | temperature | 10 | 720 | > th | dose dependent | rect. | 16 | 23 |
| 370 | 17.85 | temperature | 15 | 720 | > th | dose dependent | rect. | 12 | 23 |
| 402 | 17.87 | temperature | 10 | 720 | > th | dose dependent | rect. | 10 | 23 |
| 428 | 17.89 | temperature | 10 | 720 | > th | dose dependent | no | NA | NA |
| 448 | 17.89 | temperature | 15 | 720 | > th | dose dependent | rect. | 23 | 23 |
| 467 | 17.90 | temperature | 30 | 720 | < th | dose dependent | rect. | 22 | 22 |
| 547 | 17.95 | temperature | 15 | 720 | > th | dose dependent | rect. | 14 | 23 |
| 683 | 18.04 | temperature | 15 | 720 | > th | dose dependent | rect. | 5 | 23 |
| 703 | 18.06 | temperature | 30 | 720 | < th | dose dependent | no | NA | NA |
| 755 | 18.08 | temperature | 30 | 720 | < th | dose dependent | rect. | 2 | 23 |
| 770 | 18.09 | temperature | 30 | 720 | < th | dose dependent | rect. | 4 | 23 |
| 794 | 18.11 | temperature | 20 | 1440 | < th | dose independent | rect. | 21 | 17 |
| 825 | 18.12 | temperature | 25 | 1440 | < th | dose dependent | rect. | 8 | 23 |
| 860 | 18.14 | temperature | 20 | 1440 | > th | dose independent | rect. | 21 | 17 |
| 955 | 18.19 | temperature | 20 | 1440 | < th | dose independent | rect. | 21 | 14 |
| 973 | 18.20 | temperature | 20 | 1440 | < th | dose independent | sin | 7 | NA |