Os07g0194500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Prolyl 4-hydroxylase, alpha subunit domain containing protein.

FiT-DB / Search/ Help/ Sample detail
|
Os07g0194500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Prolyl 4-hydroxylase, alpha subunit domain containing protein.
|
|
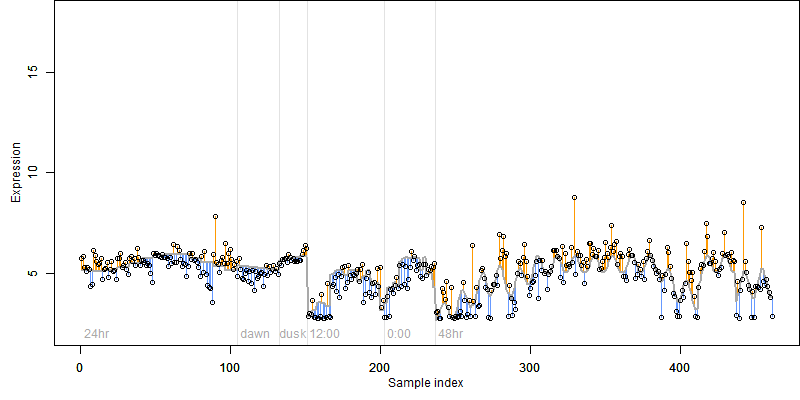
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

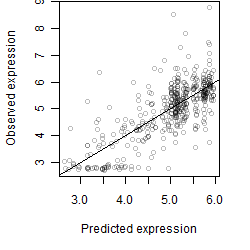 __
__
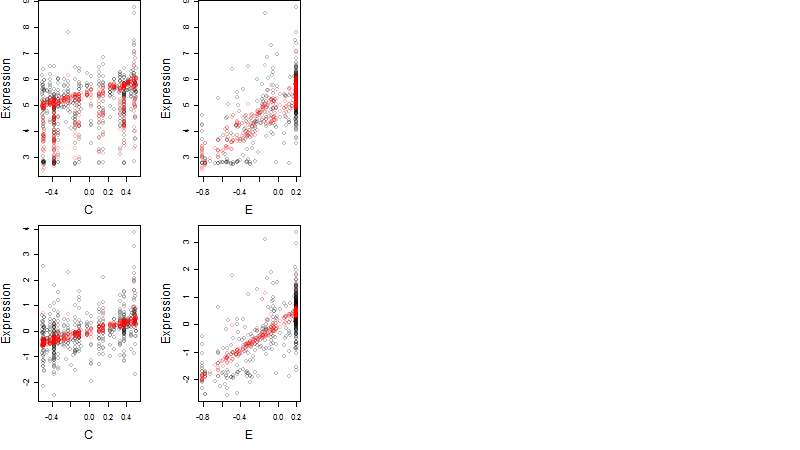
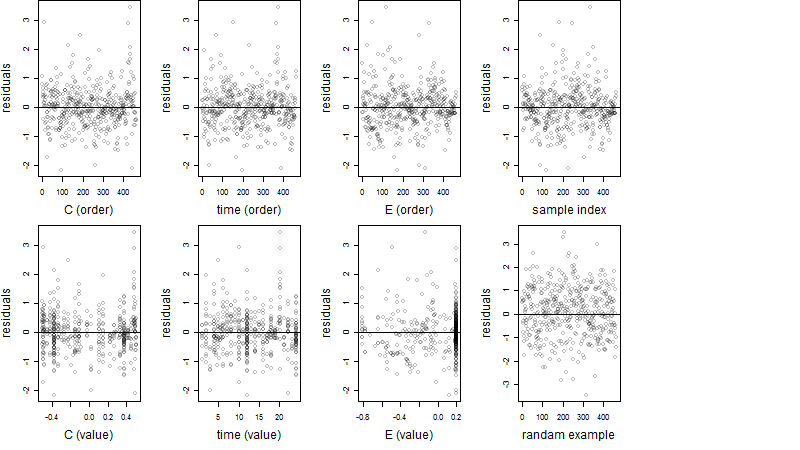
Dependence on each variable
Residual plot
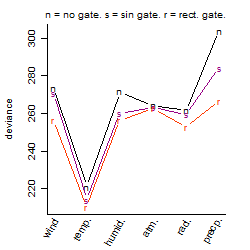
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 210.39 | 0.681 | 4.95 | 0.584 | 0.886 | 2.22 | 1.19 | 0.384 | 0.197 | 21 | 21.5 | 2230 | -- | -- |
| 210.7 | 0.676 | 4.96 | 0.482 | 0.887 | 2.2 | 1.23 | -- | 0.171 | 21 | 21.5 | 2235 | -- | -- |
| 214.96 | 0.683 | 4.94 | 0.604 | 0.878 | 2.24 | -- | 0.609 | 0.209 | 21.2 | 21.5 | 2213 | -- | -- |
| 215.78 | 0.684 | 4.96 | 0.439 | 0.88 | 2.2 | -- | -- | 0.169 | 21.2 | 21.45 | 2229 | -- | -- |
| 237.49 | 0.718 | 4.89 | 0.907 | -- | 2.25 | -- | 1.05 | 0.293 | -- | 21.3 | 799 | -- | -- |
| 346.24 | 0.872 | 4.82 | 1.52 | 1.04 | -- | 1.22 | -- | 0.835 | 21.3 | -- | -- | -- | -- |
| 351.32 | 0.878 | 4.82 | 1.48 | 1.04 | -- | -- | -- | 0.832 | 21.5 | -- | -- | -- | -- |
| 242.76 | 0.726 | 4.92 | 0.7 | -- | 2.2 | -- | -- | 0.227 | -- | 21.4 | 813 | -- | -- |
| 219.95 | 0.691 | 4.98 | -- | 0.87 | 2.36 | -- | -- | 0.0813 | 21.2 | 21.4 | 2246 | -- | -- |
| 414.47 | 0.951 | 4.79 | 1.48 | -- | -- | -- | -- | 0.829 | -- | -- | -- | -- | -- |
| 408.83 | 0.946 | 4.85 | -- | 1.04 | -- | -- | -- | 0.667 | 21.6 | -- | -- | -- | -- |
| 253.66 | 0.742 | 4.95 | -- | -- | 2.36 | -- | -- | 0.112 | -- | 21.37 | 830 | -- | -- |
| 472.6 | 1.01 | 4.83 | -- | -- | -- | -- | -- | 0.664 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance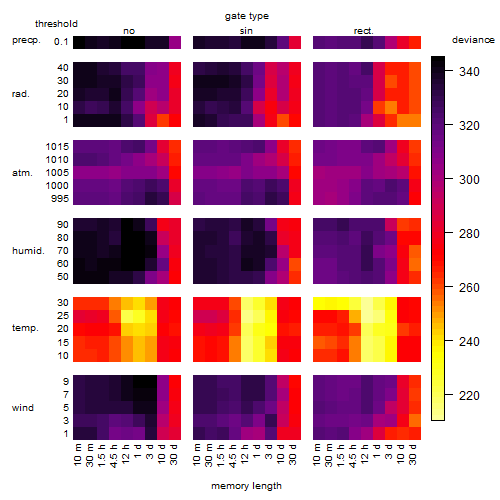
|
Histogram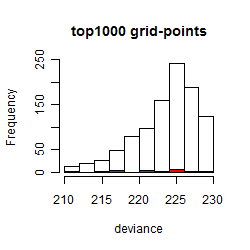 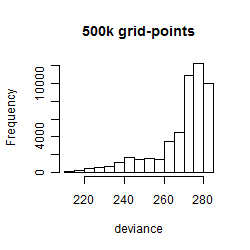
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 210.20 | temperature | 25 | 720 | < th | dose dependent | rect. | 20 | 13 |
| 8 | 211.26 | temperature | 25 | 720 | < th | dose dependent | rect. | 20 | 18 |
| 34 | 214.04 | temperature | 25 | 720 | < th | dose dependent | sin | 9 | NA |
| 58 | 215.86 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 2 |
| 64 | 216.30 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 4 |
| 70 | 216.70 | temperature | 15 | 720 | > th | dose dependent | rect. | 19 | 16 |
| 92 | 217.53 | temperature | 25 | 1440 | < th | dose dependent | rect. | 19 | 9 |
| 98 | 217.69 | temperature | 15 | 720 | > th | dose dependent | sin | 9 | NA |
| 213 | 220.63 | temperature | 25 | 720 | < th | dose dependent | rect. | 1 | 23 |
| 220 | 220.77 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 241 | 221.27 | temperature | 15 | 1440 | > th | dose dependent | rect. | 20 | 10 |
| 288 | 222.03 | temperature | 25 | 720 | < th | dose dependent | rect. | 4 | 23 |
| 442 | 223.96 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 1 |
| 511 | 224.67 | temperature | 15 | 1440 | > th | dose dependent | rect. | 1 | 6 |
| 533 | 224.84 | temperature | 15 | 1440 | > th | dose dependent | rect. | 23 | 1 |
| 541 | 224.92 | temperature | 25 | 720 | < th | dose dependent | rect. | 6 | 23 |
| 615 | 225.46 | temperature | 15 | 1440 | > th | dose dependent | rect. | 3 | 1 |
| 634 | 225.60 | temperature | 10 | 1440 | > th | dose dependent | rect. | 5 | 1 |
| 697 | 226.07 | temperature | 30 | 1440 | < th | dose dependent | rect. | 5 | 1 |
| 935 | 228.85 | temperature | 20 | 1440 | > th | dose independent | rect. | 20 | 12 |
| 1000 | 229.61 | temperature | 20 | 4320 | > th | dose independent | rect. | 21 | 2 |